The increasing global incidence and more severe clinical sequelae have arisen due to genetic modifications that confer antibiotic resistance. The resulting emergence of hypervirulent C. difficile strains, for example, NAP/BI/027, has caused outbreaks with increased mortality and morbidity worldwide [8].
In the Kingdom of Saudi Arabia (KSA), the prevalence of CDI remains unknown. However, a recent study published from the KSA used enzyme immuno-assays to determine that the incidence of CDI was 4.8% in 2007 and 4.2% in 2008; additionally, the hypervirulent NAP/BI/027 strain was identified in a case report [9,10]. These observations have raised the following questions. Is the incidence of CDI particularly low in the KSA or is it just not being detected (i.e., under-ascertainment)? Is the test used for diagnosis efficient and sufficient to inform treatment approaches that minimise the spread of infection? Which strains are circulating among patients and which antibiotics have been rendered ineffective?
The present study aimed to ascertain whether there has been an increase in the reported incidence of CDI due to advancements in diagnostic technology. Therefore, this study establishes the prevalence of C.difficile in patients with IBD from four major hospitals in the Eastern Province of the KSA, compares two methods of CDI determination, and identifies the genotype-based antimicrobial resistant pattern of the isolated C.difficile strains.
Materials and Methods
This was a prospective study conducted over a period of 14 months from October 2015 to May 2016 at the King Fahad Hospital of the University (KFHU) in Al Khobar, King Fahad Specialist Hospital (KFSH) in Dammam, Dammam General Hospital (DGH) in Dammam and Qatif Central Hospital (QCH) in Qatif. The inclusion criteria were as follows: physician evaluation and the presentation of symptoms in accordance with the international classification of diseases code ICD-10-CM- A04.7 i.e., “episode of CDI that occurs eight weeks after the onset of a previous episode, provided the symptoms from the previous episode have resolved [11]. Stool samples were collected from patients with ulcerative colitis or Crohn’s disease who attended the gastrointestinal clinic for follow-up. Infants under two years old were excluded from participation in this study.
Ethical Considerations
Prior to participation in the study, informed consent was obtained from every patient by the principal investigator and included a description of the research title, study objectives, risks related to participation, and rights of the participants. Ethical approvals were obtained from all four hospitals’ Institutional Review Boards (IRBs): Ministry of Health of King Fahad Medical City in Riyadh (#15-320E); Qatif Central Hospital (#QCHR0034); Imam Abdulrahman Bin Faisal University (IRB-PGS-2015-03-169); and King Fahad Specialist Hospital (EXT0314). All procedures were conducted in accordance with the Declaration of Helsinki.
Sample Processing and Analysis
The collected samples (374) were processed either in the Microbiology Laboratory, King Fahad Hospital or in the Microbiology Research Laboratory, Department of Clinical Laboratory Science, Imam Abdulrahman Bin Faisal University. The stool samples were either processed immediately or stored at -20°C until processed. The sample was mixed with thioglycollate broth and placed in a water bath at 80°C for 10 minutes [12]. All samples were handled with caution and processed in a Biosafety Level 2 (BSL-2) cabinet in accordance with safety guidelines and while wearing the appropriate personal protective equipment.
Glutamate Dehydrogenase (GDH) and C. difficile Toxin Testing
Samples over 1 g were divided into two portions for the GDH and toxin production assays. All of the samples were analysed using quantitative enzyme immunoassays according to the manufacturer’s instructions {VIDASkit (GDH and toxin), Biomerieux, France}.
C. difficile Culture and Identification
The stool samples were cultured on chromogenic agar (Chromagar, France) and incubated for up to 48-hours in an anaerobic jar using anaerobic pouch and incubated for 48-hours at 37°C. The culture plates were exposed to ultraviolet light at 365 nm to observe the fluorescent colonies of C. difficile. At least 3 to 5 fluorescent C. difficile colonies were isolated and stored at -80°C freezer in a Cryobank vials containing a cryogenic solution and freezable beads (Fisher Scientific, USA) until further analysis. C. difficile identification was further verified and confirmed by using the VITEK 2 ANC ID system in the Microbiology Laboratory, KFUH.
Antimicrobial Suseptibilty Testing
Antibiotic susceptibility testing was performed using E-test (BioMérieux, Craponne, France) and breakpoints [13]. The Minimal Inhibitory Concentrations (MIC) breakpoints of E-tet were used as follows: Moxifloxacin (MX) Susceptible (S), ≤2 μg/mL; moxifloxacin Resistant (R), >2 μg/mL; Vancomycin (VA) susceptible, ≤2 μg/mL; vancomycin resistant, >2 μg/mL; metronidazole (MZ) susceptible, ≤4 μg/mL; metronidazole resistant, >4 μg/mL. The following control strains were used: Clostridium difficile (ATCC700057) and Pseudomonas aeruginosa (ATCC27853).
DNA Extraction
Isolated C. difficile colonies were suspended in 300 μL of deionised water and boiled for 20-minutes at 95°C in a water bath. The samples were then incubated for 15 minutes in an ultrasonic bath and the extracted DNA in supernatant of sample after sonication was stored at -20°C until future use.
Molecular Analysis
Molecular assay was evaluated by using the new PCR based C. difficile GenoType C Diff kit assay (Hain Lifesciences, Nehren, Germany). The GenoType CDiff assay detects two C. difficile-specific genes (tpi and an undisclosed target), all known C. difficile toxins genes (tcdA, tcdB, cdtA and cdtB), the highly pathogen and virulent ribotypes 078, 126 and 027, three different deletions in the tcdC gene (the 18 bp and 39 bp deletions and the deletion at position 117) and two different mutations in the gyrA gene that have been previously associated with resistance to moxifloxacin. The GenoType C Diff assay detection is done in a line probe format (DNA-strip) and was performed according to the manufacturer’s instructions. Extracted genomic DNA from C. difficile isolate (5 μL) was used as template in a PCR reaction total volume of 50 μL and containing: 35 μL primer nucleotide mix (Hain Lifesciences, Nehren, Germany), 0.2 μL Taq DNA polymerase (Qiagen, Germany), 5 μL 10x PCR buffer, 2 μL 25 mM MgCl2 (Qiagen, Germany) and 2.8 μL deionised water (Promega, USA). PCR cycling conditions were as follows: five minutes at 95°C for one cycle, followed by 30 seconds at 95°C for 10 cycles and 58°C for two minutes for 10 cycles. Then followed by 30 cycles of 95°C for 25 seconds, 53°C for 40 seconds and 70°C for 40 seconds and finally 70°C for eight minutes. All samples amplification products were hybridised to assay strips according to the manufacturer’s instructions with incubations carried out using a Twin Cubator incubator (Hain Lifesciences, Nehren, Germany). Tested samples showing a positive result for both the ‘C diff’ and ‘tpi’ loci and at least one of the toxin genes were recorded as positive according to the manufacturer’s instructions.
Results
Sample Sites and Clinical Information
A total of 374 samples were collected from four major hospitals in the Eastern Province of the KSA. The majority of the samples were obtained from King Fahad Specialist Hospital in Dammam (42%, n=157), followed by King Fahad University Hospital in Al Khobar (35%, n=133), Qatif Central Hospital in Qatif (12%, n=44), and Dammam General Hospital in Dammam (11%, n=40) [Table/Fig-1]. The mean (±SD) age of the patients was 38 (±22.6) years and ranged from 1 to 101 years. The percentage of males was 50.3% (n=188) and 49.7% (n=186) were females.
Sample source and numbers.
| Sample source | Bed capacity | Sample numbers |
|---|
| KFHU | 500 | 133 |
| KFSH-D | 633 | 157 |
| GCH | 355 | 44 |
| DGH | 400 | 40 |
| Total | 1,888 | 374 |
KFHU: King Fahd Hospital of the University; KFSH-D: King Fahad Specialist Hospital-Dammam; QCH: Qatif Central Hospital; DGH: Dammam and Qatif Central Hospital
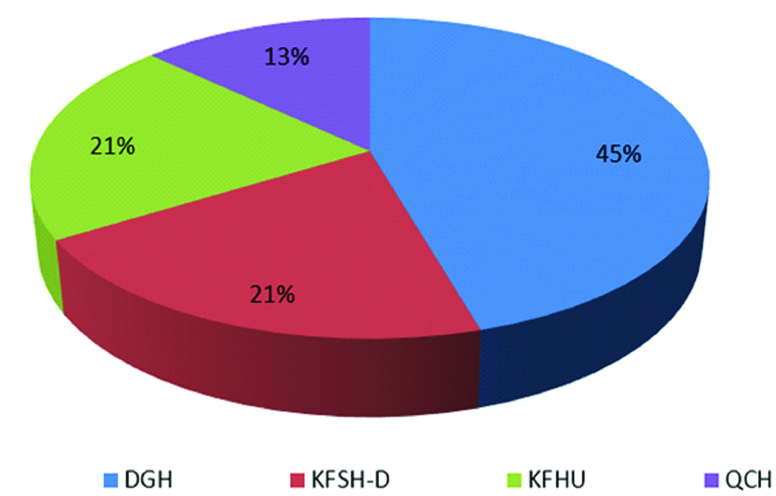
A total of 77 patients with IBD were included in the study. The mean age (±SD) of these affected individuals was 31 (±16.5) years and ranged from 7 to 82 years. 45% (n=35) were male and 55% (n=42) were female. The majority of these samples came from DGH (45.45%, n=35), followed by KFHU and KFSH (20.7%, n=16 for both), and QCH (12.98%, n=10) [Table/Fig-2].
Distribution of patients with inflammatory bowel disease by hospital.
Distribution and Prevalence of C. difficile
Of the 374 analysed samples, only 88 yielded a positive culture for C. difficile. In these infected patients, the mean (±SD) age was 31 (±22.78) years and ranged from 1 to 101 years. 51.1% (n=45) were male and 48.9% (n=43) were female. The majority of the samples were from KFSH (47%, n=41), followed by KFHU (35%, n=31), Qatif Central Hospital (9%, n=8) and DGH (9%, n=8). The GDH toxin test was positive in 82 (93.2%) of the 88 isolated C. difficile strains. Of these 82 strains, 20 (24.4%) were positive for toxin. The overall prevalence rate of C. difficile in the Eastern Province was 88 (23.5%) out of the 374 stool samples screened in this study [Table/Fig-3]. The highest number of positive samples and toxigenic strains were collected from KFSH in Dammam and KFHU in Al Khobar as presented in [Table/Fig-3].
Overall prevalence of C. difficile.
| Location | Hospital | Number of samples | Number of positive samples for C. difficile (%) | Number of non-pathogenic strains (%) | Number of toxigenic strains (%) |
|---|
| AL Khobar | KFUH | 133 | 31 (23.3) | 6 (4.5%) | 25 (18.7%) |
| Dammam | KFSH | 157 | 41 (26.1) | 9 (5.7%) | 32 (20.3%) |
| Qatif | QCH | 44 | 8 (18.2) | 1 (2.3%) | 7 (15.9%) |
| Dammam | DGH | 40 | 8 (20) | 2 (5%) | 6 (15%) |
| Total | 374 | 88 (23.5) | 18 (4.8) | 70 (18.7%) |
Antibiotic Susceptibility
The majority of the C. difficile strains were sensitive to vancomycin (96.6%, n=85), moxifloxacin (97.7%, n=86) and metronidazole (96.6%, n=85). Only three strains (3.4%) were resistant against vancomycin and metronidazole and two strains (2.3%) were resistant to moxifloxacin as shown in [Table/Fig-4].
Antibiotic susceptibility of C. difficile (n=88).
| Antibiotic | MIC breakpoint/μg/mL | Frequency (n) | Percent (%) |
|---|
| Moxifloxacin | S≤2 | 86 | 97.7 |
| R>2 | 2 | 2.3 |
| Vancomycin | S≤2 | 85 | 96.6 |
| R>2 | 3 | 3.4 |
| Metronidazole | S≤4 | 85 | 96.6 |
| R>4 | 3 | 3.4 |
Genotype and Toxigenic Profile Prevalence
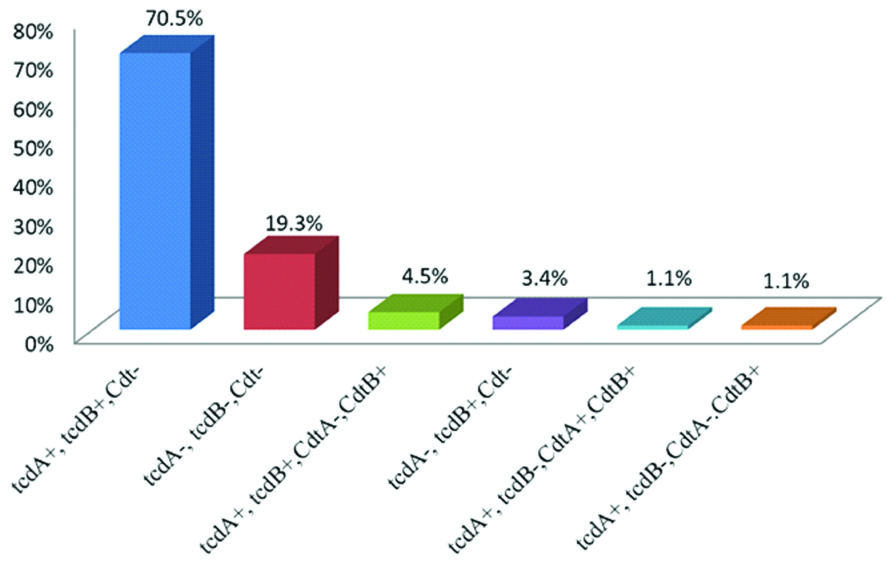
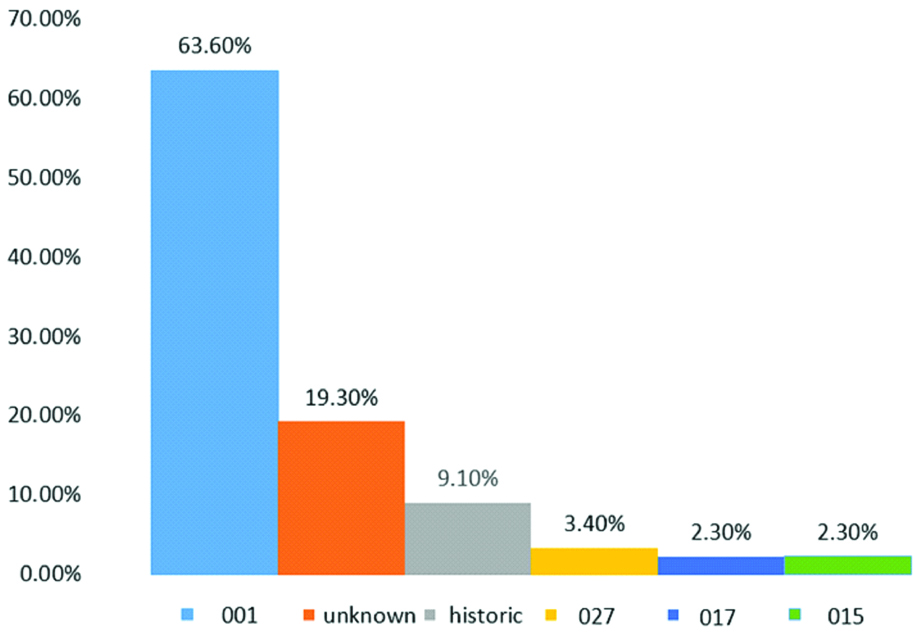
Toxigenic profile assays revealed that the majority of the strains (70.5%, n=62) were tcdA+, tcdB+, Cdt-, followed by tcdA-, tcdB-, Cdt- (19.3%, n=17), tcdA+, tcdB+, CdtA-, CdtB+ (4.5%, n=4), tcdA-, tcdB+, Cdt- (3.4%, n=3), tcdA+, tcdB-, CdtA+, CdtB+ (1.1%, n=1), and tcdA+, tcdB-, CdtA-, CdtB+ (1.1%, n=1) [Table/Fig-5]. The prevalence of genotype 001 was 63.6% (n=56), followed by 19.3% (n=17) with non-identified genotypes, 9.1% (n=8) with unknown genotypes, 3.4% (n=3) were historic 027, and 2.3% (n=2) were 017 and 015 [Table/Fig-6]. This study revealed that KFUH, KFSH-D and QCH were more likely to have the 001 genotype and its prevalence exceeded 50%. However, at QCH, the prevalence of 001 was equal to the non-pathogenic strain (both 25%). The hypervirulent ribotype historic 027 was found in KFUH (3.2%) and QCH (12.5%). Other genotypes included 015 (QCH, prevalence of 12.5%), 017 (KFSH-D, prevalence of 4.9% and DGH, prevalence of 12.5%). However, 9.1% of the strains were of unknown genotype and further tests are required to identify these.
Toxigenic profile prevalence of C. difficile.
Genotype prevalence of C. difficile.
Genotype and Toxigenic Profile Prevalence in Patients with IBD
Of the 77 patients with IBD, positive cultures were found in 22 patients (28.5%), of which 19 were toxigenic (86.4%) and three were non-pathogenic (13.6%). The most common toxigenic profile was tcdA+, tcdB+, Cdt- (89.5%) and two strains were tcdA-, tcdB+, Cdt- (10.5%). The distribution was as follows: n=6(100%) in KFSH-D where the genotype was 001 and the toxigenic profile was tcdA+, tcdB+, Cdt-. In KFUH (n=6), five (83%) were 001 genotype and tcdA+, tcdB+, Cdt- was the toxigenic profile. One (17%) was an unknown genotype with the tcdA-, tcdB+, Cdt- toxigenic profile. In DGH, six were toxigenic, of which 3 (50%) had an unknown genotype with the tcdA+, tcdB+, Cdt- toxigenic profile, 2 (33%) were 001 tcdA+, tcdB+, Cdt- and one (17%) was 017 with tcdA-, tcdB+, Cdt-. In QCH, 1 (100%) was 001 and tcdA+, tcdB+, Cdt- was the toxigenic profile. The predominant genotype among the patients with IBD from KFSH-D and QCH was 001 (100%). In KFUH, 83% were 001 and 17% had an unknown genotype, while in DGH, 33% were 001, 17% were 017 and 50% were unknown.
Discussion
The increasing incidence of CDI represents a threat to public health in general and healthcare facilities specifically [4]. Accurate testing is necessary for effective diagnosis, treatment and prevalence determination. In this study, C. difficile strains were successfully isolated from 88 out of 374 human faecal samples obtained from four different hospitals in the Eastern Province of the KSA. Several studies and international organisations {e.g., the Society for Healthcare Epidemiology of America (SHEA), the Infectious Diseases Society of America (IDSA)} have demonstrated that GDH can be reliably used as a screening tool for C. difficile in faecal samples [14,15]. In this study, the GDH test was performed using the VIDAS system and compared to CHROM agar cultures. Authors found that 95.4% of the screened samples were negative for GDH using the culture-based approach. However, while a negative GDH result can eliminate CDI from the potential diagnoses, a positive GDH result is not a conclusive indicator of CDI. The toxin assay should also be performed after positive GDH results to confirm the presence of CDI [15]. From these two tests, authors estimated that the overall prevalence of CDI in the Eastern Province was 23.5% and the overall prevalence of toxigenic C. difficile was 18.7%, which is in agreement with published international reports (range 15-25%) [16].
The main determinant of virulence in C. difficile is the presence of toxins, regardless of whether they are toxin A or B. In this study, authors performed the VIDAS CDAB test and compared these results with those from the GenoType C Diff test, which is based on PCR reverse hybridisation gene detection. Unfortunately, only 25% of the VIDAS CDAB results matched the GenoType C Diff test results (low sensitivity), which is similar to other published studies, though some of these demonstrated that the sensitivity reached 76% and 45% [17,18]. These data suggest that some CDI cases might be missed using the VIDAS CDAB assay. The prevalence of CDI in patients with IBD was 28% and their toxigenic type was tcdA+, tcdB+, Cdt-, which is consistent with recent research in China that described a prevalence rate of 32% [19]. In contrast, the reported incidence of CDI among patients with IBD was 0.4% in 2012-2013 according to the European Crohn’s and Colitis Organisation [20]. In a Romanian study, 33.3% of patients with IBD were infected with C.difficile and this is in line with the results of the present study. However, because the present study did not include clinical data, authors could not differentiate between Crohn’s disease and ulcerative colitis [21].
In order to gain insight into the molecular epidemiology of the C. difficile strains found in the Eastern Province of Saudi Arabia, GenoType C Diff assay was performed, which contains DNA probes for toxin genes A and B (tcdA and tcdB) and binary toxin genes Cdt (cdtA and cdtB). In addition, this test detects deletions in the regulatory gene tcdC and specifies the genotype (tcdC genotyping) while also being able to detect the presence of the most common mutations in gyrA (gyrA MUT1A, gyrA MUT1B). The present study revealed that the majority (70.5%, 62/70) of the toxigenic strains were toxins A and B positive and that six of the strains were binary toxin positive. The presence of either one of these genes is associated with a high incidence of recurrent infection and a high mortality rate [22]. These findings are in agreement with published reports from China, Iran and Canada [18,23,24]. However, in Europe, 6.2% of toxigenic strains were tcdA-, tcdB+, which starkly contrasts the results in this study [25]. However, in Europe, the prevalence of tcdA-, tcdB+ was higher than in the present jurisdiction but lower than in Canada. Moreover, the overall prevalence of A+B+Cdt+C. difficile strains, which are hypervirulent and associated with high mortality rates, was much lower than in North America and Europe [22].
In this study, authors evaluated three antibacterial agents two of which are currently used as standard treatment for CDI, VA and MZ. A total of 85 out of 88 C. difficile strains were sensitive to VA and MZ (both 96.6%). There were three strains that exhibited resistance to VA (3.4%) and MZ (3.4%); these were ribotype 017, a non-pathogenic ribotype, and an unknown ribotype. In a recent study in Israel, resistance to metronidazole and/or vancomycin was found in 4 out of 7 strains capable of causing re-infection [26]. Because clinical information was not included in this study, authors could not determine whether the resistant strains were from patients infected for the first time or from cases of re-infection. Two ribotype 015 strains exhibited resistance to moxifloxacin (2.3%) and the remaining 86 were sensitive to this agent. A North American study in 2012 found that over 90% of ribotype 027 strains were resistant to moxifloxacin; this contradicts the results of the present study, in which all of the ribotype 027 strains were sensitive to moxifloxacin [27]. A recently published study 2016 from Kuwait reported that the rate of resistance to metronidazole was 2.9%, similar to what authors found in this study; however, no vancomycin-resistant strains were observed in the Kuwait study [28]. In the present study, the prevalence of vancomycin resistance was 3.3%, higher than the 0.9% reported in a surveillance study conducted across 22 European countries in 2015 [29].
Authors also examined the molecular epidemiology of the C. difficile strains found in the Eastern Province of the KSA. The Genotype C. Diff assay was used because, according to the National Reference Laboratory for C. difficile, Saint Antoine Hospital AP-HP, Paris, France, this test is both rapid and accurate [30]. To determine the ribotype of the strains isolated in this study, authors examined deletions in the regulator gene tcdC. The most prevalent ribotype was 001 (63.6%), followed by unknown genotypes (9.1%), historic 027 (2.3%), 017 (2.3%) and 015 (2.3%). The dominant ribotype 001 is the same ribotype present in some European countries. In Germany, 55% of toxigenic C.difficile cases were identified as ribotype 001, which aligns with the present findings [31-33]. In Kuwait, the most common ribotype was 139 [34]. From a review of studies from Asian countries, the predominant ribotype in China was 017 and the predominant ribotype in Japan was 018 [35].
Limitation
Insufficiency of some clinical data in hospitals with regard to the antibiotic administered for primary, recurrent infection and Inflammatory Bowel Disease (IBD) such as Crohn’s or ulcerative colitis, effecting the sensitivity of the study.
Conclusion
The high prevalence of toxigenic strains described here indicates that CDI maybe an underestimated public health concern in the Eastern Province of the KSA. Genotype 001 is the predominant strain of C. difficile present in Eastern Province of KSA. Vancomycin and metronidazole resistant strains were encountered. GDH testing is the first step of workflow in limited resources hospital. A negative GDH can eliminate the diagnosis for CDI but a positive GDH is not a conclusive test for CDI. To the best of authors’s knowledge, this is the first study to describe the genotypes and toxigenic profiles of C. difficile and their respective prevalence in the Eastern Province of the KSA.