Urinary tract infection represents the most frequent bacterial infection encountered in hospital settings with an estimated prevalence of 42% [1]. Globally, symptomatic UTIs result in more than five million visits to outpatient clinics and nearly 100,000 hospitalisations annually [1].
Non-fermenting Gram-negative bacilli are a diverse group of organisms that are incapable of utilizing carbohydrates as a source of energy. They were considered as commensals or incidental organisms; however, recently they had emerged as a “nightmare bacteria” as they are associated with life-threatening infections such as pneumonia, meningitis, UTI and septicaemia [1]. Pseudomonas spp., Acinetobacter spp., Stenotrophomonas maltophilia, Burkholderia cepacia and Sphingobacterium spp., are the most clinically significant pathogens among NFGNB [2].
Recently NFGNB are being increasingly detected in significant bacteriuria and account for nearly ≥25% of hospital-acquired UTIs [3]. They are generally MDR, with an increase in resistance to cephalosporins and carbapenems. Resistance not only compromises treatment but also leads to increased mortality, and inflated cost in hospitals [3].
Carbapenems are considered as a backbone for salvage treatment and antibiotics of last resort against infections caused by MDR Enterobacteriaceae and NFGNB. However, antibiotic treatment of UTI is becoming increasingly difficult due to emerging resistance through the spread of carbapenemases leaving narrow therapeutic options [4].
Increasing alarm of CRNFs is worrisome as their genes have rapid mobility across other species and remarkable ability to hydrolyze virtually all drugs in that class [5].
The era of MDR NFGNB has renewed interest in old forgotten antibiotics such as polymyxins and fosfomycin as they have made powerful and excellent entry. Fosfomycin is a broad-spectrum antibiotic available for oral use and is recommended as one of the first line therapies for uncomplicated UTI [6].
Unfortunately, there is paucity and limited data about CRNFs isolated from patients with UTI and about the role of fosfomycin as a promising alternative therapy in CRNFs UTI [6].
The aim of this study was to determine the incidence of NFGNB in hospital-acquired UTI and to ascertain the associated risk factors. Clinical usefulness of different phenotypic and molecular tests for carbapenemases detection was also evaluated. It becomes now imperative to know about the role of fosfomycin in UTI therapy.
Materials and Methods
This cross-sectional study was conducted over a period of 18 months (from January 2017 to July 2018) during which 403 patients (180 males and 223 females) who developed symptomatic UTI at least 48h after admission to different departments of Menoufia University Hospital (MUHs) were enrolled in this study. Their ages ranged from 5 to 70 years. A written informed consent was obtained from each patient or from their guardians before inclusion in the study. Medical and demographic data of each patient was obtained. The study protocol was approved by the local ethics committee of the Menoufia University (No.2216/15-11-2016).
Urine specimens (mid-stream and catheter catch) were taken to the Microbiology laboratory, Faculty of Medicine, Menoufia University where it was examined microscopically for pus cells. Quantitative urine cultures were immediately performed on MacConkey’s and blood agar plates (Oxoid; England) using a 0.001 mL calibrated loop. Plates were incubated for 48 h at 37°C. Isolation and identification of the urinary pathogens were done according to the standard bacteriological techniques [7]. Species were identified using the Vitek-2 system (bioMerieux, France). Acinetobacter spp. and P.aeruginosa isolates were collected and subjected to further processing as follows:
Antimicrobial Susceptibility Testing
This was performed by disk diffusion method against different antibiotics (Oxoid); Zone diameters were interpreted as per CLSI, (2017) [8]. Due to the lack of acknowledged fosfomycin breakpoints for bacteria other than E. coli and E. faecalis [9] in the CLSI guidelines’, the current study applied the fosfomycin breakpoints for E. coli as per the CLSI (2017) [8] for both Acinetobacter spp. and P. aeruginosa isolates, a practice that was also followed by other authors of similar studies [10]. The isolate is susceptible if there is a zone of inhibition ≥16 mm, intermediate with a zone of 13 to 15 mm and resistant if a zone of ≤12 mm is found around fosfomycin 200 μg disk [11].
Different Phenotypic Tests for Demonstration of Carbapenemase Producers among Carbapenem-Resistant Acinetobacter and P.aeruginosa Uropathogens
Modified Carbapenem Inactivation Method (mCIM): One μL loopful of test isolate from an overnight agar plate was transferred to a tube containing 2 mL of Trypticase Soy Broth (TSB; Oxoid). The suspension is vortexed and then a meropenem disk (10 μg; Oxoid) is added to the suspension and incubated for 4 h at 35°C. Just prior to completion of the 4h incubation cycle, a 0.5 McFarland suspension of E. coli (ATCC 25922; QC strain) is inoculated onto a Muller Hinton agar plate (MHA). The meropenem disk is then removed from the TSB suspension and immediately placed on the MHA plate that has been inoculated with the QC strain. The plate is incubated at 35°C. Following over night incubation, the zone of inhibition around the meropenem disk is measured and the results are interpreted as per (CLSI, 2017 update) as follows; the isolate is considered carbapenemase producer if there is a zone of 6-15 mm and/or presence of colonies within a 16-18 mm zone around meropenem disk. However, the presence of a zone of inhibition ≥19 mm is interpreted as negative carbapenemase producer [12].
Modified Carpa NP test (CNPt-direct): Depends on a modified protocol that allows direct usage of bacterial colonies (instead of bacterial extraction buffer used in the original Carpa NP test) to provide simplicity and cost reduction per reaction as follows:
A full 1 μL loop of a pure bacterial culture recovered from MHA plate was directly suspended in 1.5 mL Eppendorf tubes containing 100 μL of CNPt-direct mix, supplemented with 12 mg/mL imipenem-cilastatin injectable form (test tube: tube B) or without antibiotic (control tube: tube A). Tubes were vigorously mixed for 5-10 sec using a vortex device before incubation. Finally, the tubes were incubated at 35°C and monitored throughout 2 h for development of color change in the antibiotic-containing tube. The previously mentioned CNPt-direct mix solution consisted of 0.05% phenol red with 0.1 mmol/liter ZnSO4 adjusted to pH of 7.8. Results were interpreted as follows: (i) both tubes A and B remaining red indicated a non-carbapenemase-producing isolate; (ii) tube A red and tube B turning yellow/orange indicated a carbapenemase-producing isolate; and (iii) both tube A and B turning yellow/orange indicated a non-interpretable result [13].
CarbAcineto NP Test: In this version, a 5M NaCl solution (Sigma-Aldrich, Saint-Quentin-Fallavier, France) was used instead of the lysis buffer to avoid the buffer effect. The bacterial inoculum was also doubled from one-third to one-half of a calibrated loop (10 μL) to a full calibrated loop (2-3 loops) in order to increase the enzyme quantity. Briefly, a full calibrated loop (10 μL) of the tested strain was recovered from MHA plates and resuspended in two 1.5 mL Eppendorf tubes (A and B) containing 100 μL of 5M NaCl. In tube A (internal control), 100 μL of the CNPt-direct mix solution was added. In tube B (test tube), 100 μL of the previously prepared mix solution supplemented with 12 mg/mL imipenem-cilastatin was added. Tubes A and B were incubated at 37°C for a maximum of 2 h. The remaining of the test and results interpretation were performed as per CNPt-direct [14].
Molecular Characterisation of Class B (blaVIM-2, blaNDM and blaIMP) and Class D (blaOXA-23) Carbapenemases among Carbapenem-Resistant Acinetobacter and P.aeruginosa Uropathogens
Genetic support for carbapenemase production was investigated by PCR assay for all Acinetobacter and P.aeruginosa uropathogens exhibiting carbapenemase-producing phenotypes. Target genes (blaVIM-2, blaIMP, blaNDM and blaOXA-23) used in the study are shown in [Table/Fig-1] [15]. The DNA from an overnight grown culture was extracted using DNA extraction kit (Qiagen-Germany) according to the manufacturer’s instructions. The final volume of the reaction mix was 25 μL (PCR Master Mix 2x, Thermo Scientific) containing 1 μL of the target DNA. Amplification for screening was carried out using GoTaq Green Master Mix (Promega) and the following thermal cycling protocol: 10 min at 94°C and 36 cycles of amplification consisting of 30s at 94°C, 40s at 52°C and 50s at 72°C, with 5 min at 72°C for the final extension step [16]. The PCR products were detected by using agarose gel electrophoresis.
Target genes used in the study [15].
| Target genes | Sequence (5’-3’) | PCR Products | Reference |
|---|
| bla VIM-2 | F-GATGGTGTTTGGTCGCATAR-CGAATGCGCAGCACCAG | 390 bp | [15] |
| bla NDM | F-GGCGGAATGGCTCATCACGAR-CGCAAC ACAGCCTGACTTTC | 621 bp |
| bla IMP | F-CTACCGCAGCAGAGTCT TTGR-AACCAGTTTTGCCTTACCAT | 232 bp |
| bla OXA-23 | F: GAT CGG ATT GGA GAA CCA GAR: ATT TCT GAC CGCATT TCC AT | 501 bp |
Statistical Analysis
Statistical analysis was done using SPSS version 20. Mean median and chi-square (χ2) was used. Statistical significance was set at p-value <0.05. Accuracy was represented using the terms sensitivity and specificity.
Results
In one and a half year duration, a total of 403 urine samples; collected from nosocomial UTI patients (180 males and 223 females) were processed for quantitative culture, all showed significant monomicrobial growth. Enterobacteriaceae were the predominant uropathogens 280/403 (69.5%). While NFGNB accounted for an isolation rate of 23.75% (96/403) of total culture positive urine samples, among them P.aeruginosa was the predominant organism; 52/96 (54.2%), followed by Acinetobacter spp.; 43/96 (44.8%) then Stenotrophomonas maltophila (only one isolate) [Table/Fig-2].
NFGN bacilli and other uropathogens clinical isolates from patients with UTIs.
| Isolates group | No. (%) | Organisms | No. (%) |
|---|
| Enterobacteriaceae | 280 (69.5%) | E. coli | 204 (72.8%) |
| Klebsiella spp. | 48 (17.1%) |
| Proteus spp. | 11 (4%) |
| Enterobacter species | 13 (5%) |
| Citrobacter spp. | 4 (1.1%) |
| NFGNB | 96 (23.75%) | P.aeruginosa | 52 (54.2%) |
| Acinetobacter spp. | 43 (44.8%) |
| Stenotrophomonas maltophila | 1 (1%) |
| Gram positive cocci | 26 (6.5 %) | Enterococcus faecalis | 8 (30.8%) |
| Staph.saprophyticus | 18 (69.2%) |
| Fungi | 1 (0.25%) | Candida albicans | 1 (100%) |
| 403 (100%) | Total | 403 (100%) |
All P.aeruginosa and Acinetobacter spp. uropathogens were MDR and showed variable susceptibilities to routinely prescribed antibiotics. The most alarming carbapenem resistance was highly exhibited by our NFGNB uropathogens; overall imipenem and meropenem resistance rate was 83.2% (79/95). P.aeruginosa and Acinetobacter uropathogens were resistant to most of the tested antibiotics with 100% resistance to piperacillin, cefotaxime, tetracycline and trimethoprim-sulfamethoxazole. Moreover, resistance to cefepime, ceftazidime and piperacillin/tazobactam exceeded 64%. Even though, all NFGNB were uniformly sensitive to colistin (100%). Also, amikacin, tobramycin and norfloxacin exhibited a remarkable high activity (>74% of isolates were sensitive) against most of P.aeruginosa and Acinetobacter uropathogens. As regards fosfomycin, higher percentages of both carbapenem-resistant P.aeruginosa (47/52; 90.4%) and Acinetobacter (38/43; 88.4%) uropathogens remained susceptible to fosfomycin when using the CLSI (2017) break points for E.coli [Table/Fig-3].
Antimicrobial susceptibility pattern of the isolated NFGNB using the disk diffusion method.
| Antimicrobial agents | P.aeruginosa (n=52) | Acinetobacter spp., (n=43) | Total (95) NFGNB |
|---|
| R (%) | S (%) | R (%) | S (%) | R (%) | S (%) |
|---|
| Piperacillin (PRL) | 52 (100%) | 0 | 43 (100%) | 0 | 95 (100%) | 0 |
| Piperacillin/tazobactam (TZP) | 38 (73%) | 14 (27%) | 23 (53.4%) | 20 (46.6%) | 61 (64.2%) | 34 (35.8%) |
| Cefotaxime (CTX) | 52 (100%) | 0 | 43 (100%) | 0 | 95 (100%) | 0 |
| Ceftazidime (CAZ) | 48 (92.3 %) | 4 (7.6%) | 39 (90.6%) | 4 (9.4%) | 87 (91.5%) | 8 (8.5%) |
| Cefepime (FEB) | 41 (78.8%) | 11 (21.2%) | 33 (76.7%) | 10 (23.3%) | 74 (77.9%) | 21 (22.1%) |
| Imipenem (IPM) | 44 (84.6%) | 8 (15.4%) | 35 (81.3%) | 8 (18.7%) | 79 (83.2%) | 16 (16.8%) |
| Meropenem (MEM) | 44 (84.6%) | 8 (15.4%) | 35 (81.3%) | 8 (18.7%) | 79 (83.2%) | 16 (16.8%) |
| Amikacin (AK) | 7 (13.5%) | 45 (84.5%) | 9 (20.9%) | 34 (79.1%) | 16 (16.8%) | 79 (83.2%) |
| Tobramycin (Tob) | 9 (17.3%) | 43 (82.7%) | 11 (25.6%) | 32 (74.4%) | 20 (21%) | 75 (79%) |
| Ciprofloxacin (CIP) | 28 (53.8%) | 24 (46.2%) | 19 (44%) | 24 (54%) | 47 (49.4%) | 48 (50.6%) |
| Levofloxacin (LEV) | 26 (50%) | 26 (50%) | 28 (65.1%) | 15 (34.9%) | 54 (56.8%) | 41 (43.2%) |
| Norfloxacin (Nor) | 11 (21.1%) | 41 (78.9%) | 13 (30.2%) | 30 (69.8%) | 24 (25.2%) | 71 (74.8%) |
| Tetracycline (TE) | 52 (100%) | 0 | 43 (100%) | 0 | 95 (100%) | 0 |
| Trimethoprim/Sulfamethoxazole (SXT) | 52 (100%) | 0 | 43 (100%) | 0 | 95 (100%) | 0 |
| Colistin (CL) | 0 | 52 (100%) | 0 | 43 (100%) | 0 | 95 (100%) |
| Fosfomycin (FF 200) | 5 (9.6%) | 47 (90.4%) | 5 (11.6%) | 38 (88.4%) | 10 (10.5%) | 85 (89.5%) |
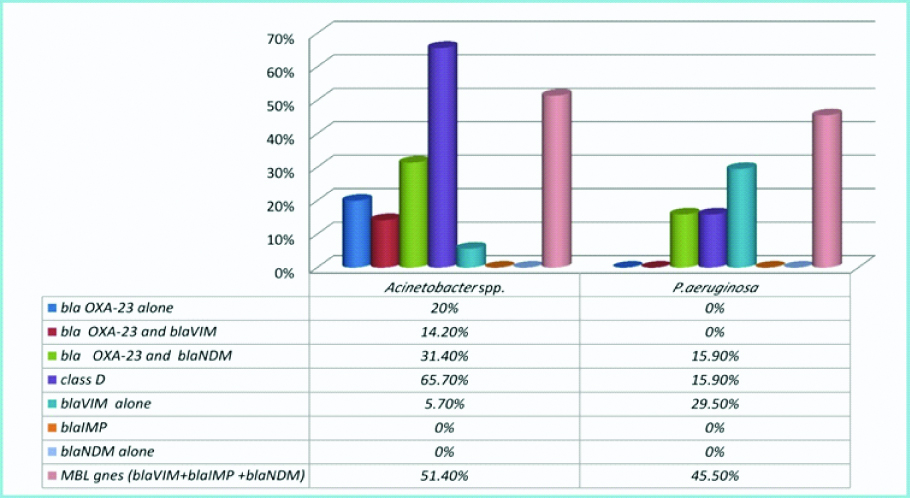
PCR results for carbapenemase genes revealed that class D and MBL genes both were represented among carbapenem-resistant NFGN uropathogens, interestingly blaNDM gene only detected in co-existence with blaOXA-23 in both P.aeruginosa (7 isolates) and Acinetobacter spp. (11 isolates). Even though, blaIMP was not detected in all NFGN clinical isolates. Carbapenem-resistant Acinetobacter spp., (25/35; 71.4%) showed high carbapenemase activity compared to P.aeruginosa (20/44; 45.5%). Carbapenemases genes harbored by Acinetobacter spp., and P.aeruginosa carbapenem-resistant uropathogens were remarkably different. Among Acinetobacter spp., uropathogens class D carbapenemase was mostly presented (23/35; 65.7%) followed by MBL genes (18/35; 51.4%). On the contrary, MBL genes were mainly presented among P.aeruginosa isolates (20/44; 45.5%) followed by class D carbapenemase (7/44; 15.9%) [Table/Fig-4,5 and 6].
Results of multiplex PCR for detection of blaVIM-2, blaIMP, blaNDM and blaOXA-23 genes among carbapenem-resistant Acinetobacter spp. and P.aeruginosa uropathogens.
| Genotypes | Carbapenem-resistant uropathogenic isolates | Total carbapenem-resistant NFGNB (n=79) |
|---|
| Acinetobacter spp. (n=35) | P.aeruginosa (n=44) | |
|---|
| No. | % | No. | % | |
|---|
| blaOXA-23 alone | 7 | (20%) | - | - | 7 (8.8%) |
| blaVIM-2 alone | 2 | (5.7%) | 13 | (29.5%) | 15 (19%) |
| blaIMP alone | - | - | - | - | - |
| blaNDM alone | - | - | - | - | - |
| Co-existence of class D +class B genes | | | | | |
| blaOXA-23 and blaVIM-2 | 5 | (14.2%) | - | - | 5 (6.3%) |
| blaOXA-23 and blaNDM | 11 | (31.4%) | 7 | (15.9%) | 18 (22.8%) |
| Total blaVIM-2 | 7 | (20%) | 13 | (29.5%) | 20 (25.3%) |
| Total blaNDM | 11 | (40%) | 7 | (15.9%) | 18 (22.8%) |
| Total class B genes (blaVIM-2 +blaNDM) | 18 | (51.4%) | 20 | (45.5%) | 38 (48%) |
| Total class D genes (bla OXA-23) | 23 | (65.7%) | 7 | (15.9%) | 30 (38%) |
| Total carbapenemase producers | 25 | (71.4%) | 20 | (45.5%) | 45 (57%) |
Multiplex PCR for detection of blaVIM-2, blaIMP, blaNDM and blaOXA-23 genes among carbapenem-resistant Acinetobacter spp. and P.aeruginosa uropathogens.
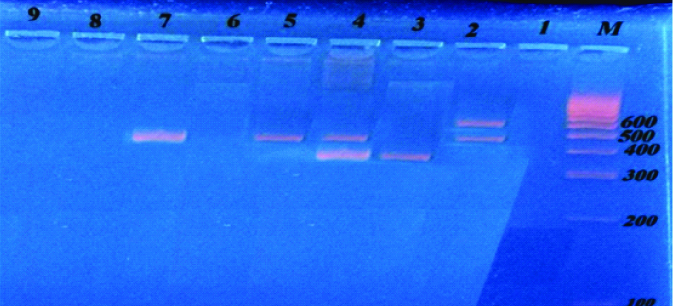
Multiplex PCR amplified products of blaOxa-23, blaVIM-2, blaIMP and blaNDM genes from Acinetobacter spp., and P.aeruginosa:
Lane M: DNA molecular size marker (100-1000 bp).
Lane 2: Positive for bla Oxa-23 (501 bp) and blaNDM (621) genes.
Lane 3: Positive for blaVIM (390bp) gene.
Lane 4: Positive for bla Oxa-23 (501 bp) and blaVIM-2 (390) genes.
Lanes 5and 7: Positive for blaOxa-23 (501 bp) gene.
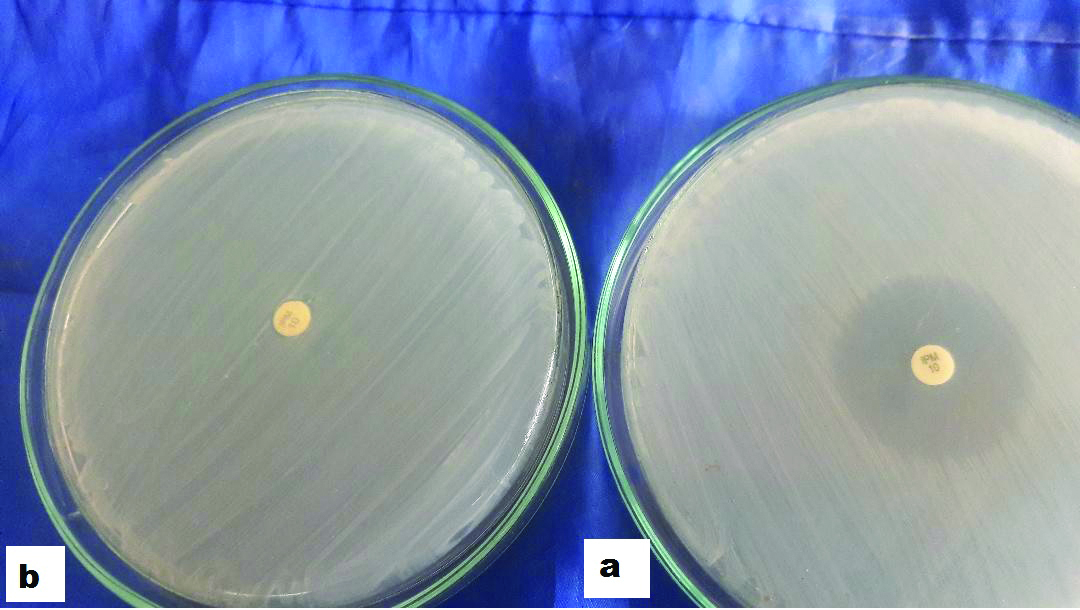
Representative results obtained from mCIM, CNPt-direct and CarbAcineto NP tests are shown in [Table/Fig-7,8]. In relation to PCR results, the sensitivity, specificity and accuracy of the mCIM were 90%, 91.7% and 90.9% respectively for P.aeruginosa when testing for class B genes and 85.7%, 91.8% and 90.9% when testing for combined class B+D genes. However, the test performed poorly with Acinetobacter spp. The CNPt-direct also achieved excellent accuracy (exceeding 90% for P.aeruginosa and ranging from 88.5% to 91.4% for Acinetobacter spp.) with all types of carbapenemases detected. For Acinetobacter spp., The CarbAcineto NP test proved 100% sensitivity with all carbapenemase classes [Table/Fig-9,10].
Representative results obtained by mCIM test; a represents negative result i.e., zone=20 mm and b represents positive result i.e., no zone of inhibition around the carbapenem disk.
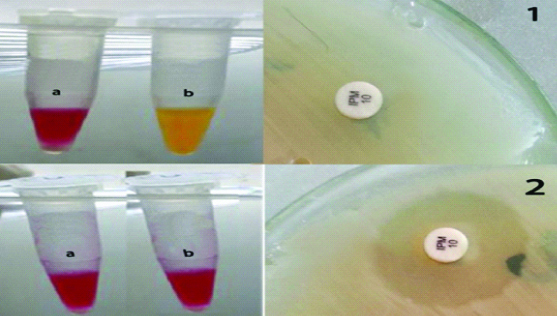
Representative results obtained by both CNPt-direct and CarbAcineto NP tests; 1 represents results obtained by imipenem- non susceptible strain where a tube (red colour/control tube) is a negative and tube b (yellow colour) is a positive result i.e., carbapenemase-producing isolate. 2 represents results obtained by imipenem-susceptible strain where both tubes (a,b) are negative results i.e., carbapenemase-non producing isolate.
Sensitivity, specificity and accuracy of three phenotypic tests in relation to PCR results of class B and class D carbapenemase detection among carbapenem-resistant Acinetobacter spp.(n=35).
| Phenotypic tests | Multiplex PCR results for blaVIM-2, blaNDM and blaOXA-23 carbapenemases in Acinetobacter spp. (n=35) |
|---|
| Ambler class B | Ambler class DblaOXA-23 alone | Combined (class B+D) |
|---|
| +ve(n=18) | -ve(n=17) | Sensitivity | Specificity | Accuracy | +ve (n=7) | -ve(n=28) | Sensitivity | Specificity | Accuracy | +ve(n=16) | -ve(n=19) | Sensitivity | Specificity | Accuracy |
|---|
| No. | % | No. | % | | | | No. | % | No. | % | | | | No. | % | No. | % | | | |
|---|
| mCIM | +ve | 5 | 27.7 | 6 | 35.3 | 27.7% | 64.7% | 45.7% | 2 | 28.6 | 17 | 60.7 | 28.6% | 39.3% | 37.1% | 5 | 31.2 | 12 | 63.2 | 31.2% | 36.8% | 34% |
| -ve | 13 | 72.3 | 11 | 64.7 | 5 | 71.4 | 11 | 39.3 | 11 | 68.8 | 7 | 36.8 |
| CNPt-direct | +ve | 16 | 88.9 | 2 | 11.8 | 88.9% | 88.2% | 88.5% | 6 | 85.7 | 2 | 7.1 | 85.7% | 92.9% | 91.4% | 14 | 87.5 | 2 | 10.5 | 87.5% | 89.5% | 88.5% |
| -ve | 2 | 11.1 | 15 | 88.2 | 1 | 14.3 | 26 | 92.9 | 2 | 12.5 | 17 | 89.5 |
| Carb-Acineto NP test | +ve | 18 | 100 | 1 | 5.9 | 100% | 94.1% | 97.1% | 7 | 100 | 1 | 3.5 | 100% | 96.5% | 97.1% | 16 | 100 | 1 | 5.2 | 100% | 94.8% | 97.1% |
| -ve | 0 | 0 | 16 | 94.1 | 0 | 0 | 27 | 96.5 | 0 | 0 | 18 | 94.8 |
Sensitivity, specificity and accuracy of two phenotypic tests in relation to PCR results of class B and class D carbapenemase detection among carbapenem-resistant P.aeruginosa isolates (n=44).
| Phenotypic tests | Multiplex PCR results for blaVIM-2, blaNDM and blaOXA-23 carbapenemases in P.aeruginosa (n=44) |
|---|
| Ambler class B | Combined (class D+B)blaOXA-23+ blaNDM |
|---|
| +ve(n=20) | -ve(n=24) | Sensitivity | Specificity | Accuracy | +ve(n=7) | -ve(n=37) | Sensitivity | Specificity | Accuracy |
|---|
| No. | % | No. | % | | | | No. | % | No. | % | | | |
|---|
| mCIM | +ve | 18 | 90 | 2 | 8.3 | 90% | 91.7% | 90.9% | 6 | 85.7 | 3 | 8.2 | 85.7% | 91.8% | 90.9% |
| -ve | 2 | 10 | 22 | 91.7 | 1 | 14.3 | 34 | 91.8 |
| CNPt-direct | +ve | 19 | 95 | 1 | 4.1 | 95% | 95.8% | 95.4% | 7 | 100 | 4 | 10.8 | 100% | 89.2% | 90.9% |
| -ve | 1 | 5 | 23 | 95.8 | 0 | 0 | 33 | 89.2 |
As regards risk factors, NFGNB UTI was more among females, although the difference was insignificant. Comparing patients infected with carbapenemase-producing NFGN uropathogens with others infected with non carbapenemase-producing isolates; catheterization ≥7 days, recurrent UTI and admission to high risk areas (ICUs, dialysis units and oncology department) were the only significant risk factors associated, also history of diabetes and associated malignancy or autoimmune disease with cortical therapy were significant factors contributing to carbapenemase-producing NFGN infections [Table/Fig-11].
Relation between carbapenemase production among the isolated NFGN uropathogens, demographic data and risk factors of the studied patients.
| Variables | Carbapenemase-producing NFGNB (n=45)No. (%) | Non carbapenemase-producing NFGNB (n=34)No. (%) | p-value |
|---|
| Age, mean years±SD | 41.4±11.8 | 36.3±10.2 | 0.067 |
| Gender | | | |
| MaleFemale | 18 (40%)27 (60%) | 14 (41%)20 (59%) | 0.998 |
| Median days of hospital stay (range) | 16 (7-28) | 12 (7-14) | 0.129 |
| No. (%) with urinary complicating factor• Catheterization <7days• Catheterization ≥7days• Recurrent UTIs • Surgery | 9 (20%)29 (64.4%)12 (26.6%)19 (42.2%) | 8 (23.5%)10 (29.4%)012 (35.2%) | 0.603<0.001**<0.001**0.822 |
| • Prior invasive procedure• Prior carbapenem administration• Prior surgery | 21 (46.7%)30 (66.7%)14 (31.1%) | 12 (35.3%)16 (47%)10 (29.4%) | 0.7220.1230.652 |
| Co-morbidities• Diabetes• Chronic liver diseases • Malignancy • Hypertension • Autoimmune disease with steroid therapy | 9 (20%)3 (6.7%)9 (20%)12 (26.6%)6 (13.3%) | 2 (5.9%)1 (2.9%)09 (26.5%)0 | 0.014*0.119< 0.001**1.000<0.001** |
| Distribution in different wardsHigh risk areas• ICUs• Dialysis units• Oncology departmentOther clinical wards | 33 (73.3%)21 (46.7%)3 (6.7%)9 (20%)12 (26.7%) | 11(31.4%)9 (25.7 %)2 (5.7%)024 (68.6%) | <0.001**<0.001** |
Discussion
Urinary tract infection ranks among the commonest infections in healthcare facilities [1]. Multi-drug resistant NFGNB, e.g., P.aeruginosa and Acinetobacter spp. are undergoing genetic modifications particularly in hospital settings where the evolution of highly resistant carbapenemases may cause untreatable UTI especially with declining number of newer antimicrobial agents entering the clinical practice [17].
In the present study, a total of 403 culture-proved UTI episodes were verified and NFGNB accounted for 23.75% (96/403) of total isolates, of which P.aeruginosa and Acinetobacter spp. were the dominant species. These results correlated with the findings of Abbas HA et al., in Egypt, Mobashshera T et al., and Jiménez-Guerra G et al., as they declared that P.aeruginosa was the most commonly isolated NFGN uropathogen followed by Acinetobacter spp. [18,19,20]. However, unusual uropathogens as B. cepacia and A. hydrophila were also reported [21]. In this study, unusual NFGNB were scarcely isolated, as variability of pathogens genera and prevalence rate in UTI has been reported [22].
Empiric antibiotic therapy is the mainstay for UTI treatment; however, this policy has increased the trend of resistance among uropathogens [22]. In the present work, the rate of MDR displayed a worrisome trend as all NFGNB uropathogens were MDR (100%), this high MDR rate was nearer to that of Abbas HA et al., and Grewal US et al., [18,22]. Even though low percentages of MDR uropathogens were reported globally; 6.9% in Canada, 35% in Germany, 20% in Malaysia and 21% in Nepal [23]. Higher resistance rate among NFGN uropathogens in our study may be explained by differences in antibiotics policy, patient’s risk factors and infection control policies between each hospital and country.
In the current study, Acinetobacter and P.aeruginosa uropathogens were found to be resistant (>75%) to most of antibiotics in current use for UTI empiric therapy including carbapenems (83.2%). High resistance rate to carbapenems, cephalosporins and quinolones, drugs that were considered effective in UTI is alarming and needs attention about antibiotic prescription policies in our country. This correlates with the results of Tohamy ST et al., and Grover N et al., [24,25]. Overuse of quinolones and cephalosporins in the last few years has contributed to high resistance to those drugs, as selective pressure is a major determinant for emergence of MDR strains [1].
Fortunately, P.aeruginosa and Acinetobacter isolates showed 100% sensitivity to colistin and more than 80% sensitivity to amikacin, this result was consistent with a previous study [26]. Also, Bader MS et al., suggested that; aminoglycosides and colistin were considered the best choice therapeutic options in UTIs associated with MDR pathogens [27].
Against a background of rapid increase of MDR Gram-negative bacilli worldwide and the resulting lack of effective antibiotic treatment, some older antibiotics have been tested for new therapeutic uses [28]. One such was fosfomycin, which according to the results of previously published studies had shown very good in vitro activity against MDR NFGNB [29,30]. All this evidence has generated higher interest in the use of fosfomycin in the last 5 years [28].
The main finding of our study was that fosfomycin exhibited considerably high antimicrobial activity against both P.aeruginosa and Acinetobacter spp., uropathogens, an observation that agreed with Perdigão-Neto LV et al., where >80% of Acinetobacter spp. and >90% of P. aeruginosa isolates were considered susceptible when applying CLSI breakpoints for E. coli [31].
For establishment of appropriate therapy and control drug resistance in UTI associated with NFGNB, molecular detection of carbapenemases encoding genes and analysis of their molecular diversity is becoming increasingly important [5].
In the present study, nearly half of carbapenem-resistant NFGN uropathogens were MBL producers which is nearer to the results of El-Mahallawy HA et al., and Grover N et al., [21,25]. However, higher rates of MBLs (75%) among NFGNB were also recorded [32]. Resistance due to MBL-encoding genes has a potential for rapid dissemination, since MBL genes are carried on highly transmissible plasmids [33]. In the present study remarkable co-existence of blaOXA-23 and blaNDM was encountered in both P.aeruginosa and Acinetobacter spp., this may explain the higher prevalence of co-resistance found in our isolates as most isolates exhibited high resistance to beta-lactams, aminoglycosides, and quinolones. Co-existence of several carbapenemases in GNB represents an issue that requires close and continuous monitoring [33].
Carbapenemase-encoding genes differ significantly among countries, so PCR was performed to identify specific profiles of uropathogens genes. In this study, Acinetobacter uropathogens exhibited high carbapenemase activity encoded by class D OXA-23 (23/35; 65.7%). This result was previously reported by Fouad M et al., Helmy OM et al., and Bouarfa N et al., [34,35,36]. Acinetobacter spp. harboring blaOXA-23 gene are becoming increasingly widespread, this oxacillinase has been extensively reported from Europe, Algeria, India, Asia [34] and Egypt [33]. blaOXA-23 oxacillinase may become the dominant and endemic gene of carbapenem resistance in Acinetobacter spp. causing UTI in Egypt hospitals. As reported, isolates harbored blaOXA-23 gene are generally MDR [34].
In agreement with a previous study in Egypt [21]; MBL genes are increasingly recognised in P. aeruginosa, in this study nearly half (20/44; 45.5%) of carbapenem-resistant P. aeruginosa were MBL producers with predominance of blaVIM-2 gene. Recently, higher prevalence of MBL genes in P.aeruginosa (>65%) has been reported [21,37]. Although only small number of isolate carried the blaNDM gene, its detection is of concern as blaNDM-producing bacteria are rapidly spreading worldwide [33].
However, several pieces of literature have reported that carbapenem resistance among P. aeruginosa strains is predominantly mediated by non-carbapenemase mechanisms, including the loss of porin expression or efflux pumps-mediated resistance [32]. That may explain other mechanisms of carbapenem resistance in non-carbapenemase producing- P.aeruginosa isolates in this study (24/44; 55%).
In this study, we attempted to identify significant risk factors for carbapenemase-producing P.aeruginosa or Acinetobacter associated UTI infection; increased duration of urinary catheterization, admission to high risk areas, history of diabetes, malignancy and autoimmune diseases were remarkable conditions that favors infection with those pathogens. Catheter disrupts host defense mechanisms and provides surface for the attachment and biofilm formation that enhance survival and decrease antibiotic action on MDR pathogens [1]. Patients who are taking corticosteroids have a propensity to develop P. aeruginosa and Acinetobacter infections. Other commonly studied risk factor (age, sex, hospital admission, prior antibiotic and surgical intervention) didn’t predict such infections with carbapenemase-producing NFGN uropathogens [25].
Notably, carbapenemase-producing NFGNB (CPNFs) represents a major threat for the medical community as the potential for dissemination is high, therefore the recent CLSI recommendations indicated that, P.aeruginosa, and Acinetobacter spp. with elevated carbapenem MICs or reduced disk diffusion inhibition zones should be tested, for the production of carbapenemases for epidemiological and infection control purposes [8].
One of the main objectives of our study was to address the most accurate phenotypic test that can be rapidly and easily applied in routine laboratories for detection of carbapenemases, as molecular methods (gold standard) may not be immediately available and may be limited by the number of targets detected [38].
As regards the mCIM test, our findings proved that the sensitivity, specificity and accuracy of that test were 90%, 91.7% and 90.9% respectively for P.aeruginosa when testing for class B carbapenemases and 85.7%, 91.8% and 90.9% for combined class B+D genes. However, for Acinetobacter spp., the test performed poorly with a sensitivity, specificity and accuracy of only 27.7%, 64.7% and 45.7% for class B genes, 28.6%, 39.3% and 37.1% for class D genes and finally 31.2%, 36.8% and 34% for combined class B+D carbapenemases. Such findings agreed with Simner PJ et al., who reported that mCIM test achieved excellent sensitivity and specificity (>90%) for detecting carbapenemase-producing P. aeruginosa isolates but not for Acinetobacter spp. possibly due to differences between P. aeruginosa and Acinetobacter in the heterogeneity of carbapenemases. The intrinsic low cell membrane permeability of Acinetobacter isolates, requiring a lysis solution different from that used for P. aeruginosa makes it more difficult for Acinetobacter carbapenemases to be detected by rapid assays that rely only on lysis and release of carbapenemases [39].
Moreover, the differences observed between P. aeruginosa and Acinetobacter may be due to the weaker carbapenem-hydrolytic activity of the class D blaOXA-carbapenemases which are the most common carbapenemases produced predominantly by Acinetobacter spp. [14,40] as proved in our study (23/35;65.7%). In the CLSI AST News Update, (2017), they also postulated that, the mCIM as described here is not reliable for detecting carbapenemase production in Acinetobacterbaumannii complex. On the contrary, the test has been shown to be reliable for detection of carbapenemase production in P. aeruginosa [11].
In this study, both the mCIM and the modified CarpaNP test (CNPt-direct) nearly achieved the same sensitivity and specificity for P.aeruginosa, however, the mCIM test is simple to perform with minimal hands-on time (<5 min per isolate when testing multiple isolates) and uses laboratory supplies that are readily available [41].
In an attempt to improve carbapenemases detection among Acinetobacter isolates, we shifted towards the CNPt-direct that provided superior results compared with the mCIM test with sensitivity of 85.7%:88.9% reaching a maximum of 100% with the CarbAcineto NP test. These results were in consistence with that reported by Bakour S et al., who stated that, the CNPt-direct method in its simple and modified protocol detected all carbapenemases with 100% sensitivity and 100% specificity. They recommended the test for detection of different carbapenemase types from P.aeruginosa and Acinetobacter spp. using a single protocol and concluded that, the effectiveness of this test on a large series of bacteria may allow us to identify the production of carbapenemases even before identification of the bacterial strain [42]. Dortet L et al., also documented that, the newly designed CarbAcineto NP test was rapid, reproducible and efficiently detected all types of carbapenem-hydrolyzing enzymes including the OXA-type carbapenemases with a sensitivity of 94.7% and a specificity of 100%. The authors confirmed that, the CarbAcineto NP is a cost-effective test that offers a reliable and affordable technique for identifying carbapenemase production in Acinetobacter spp., which is a marker of multidrug resistance in those species [14].
The better detection of blaOXA-type carbapenemase in Acinetobacter isolates when using CarbAcineto NP test is probably due to two independent factors. First, the bacterial inoculum used in the CarbAcineto NP test was doubled compared to that in the CNPt-direct, leading to an increased amount of enzyme released in the revealing solution. Second, is that the use of the hyper-osmotic 5 M NaCl solution provides efficient lysis of the bacteria, easy release of the enzymes and consequently better detection [14].
Several researches declared that, the newly performed phenotypic tests like mCIM test, the CarpaNP test with its modified protocol and the CarbAcineto NP test, have efficiently improved the detection of all types of carbapenemases particularly among the NFGNB. Such new tests succeeded to overcome most drawbacks and false-negative results obtained with other phenotypic tests e.g., The modified Hodge test and combined disk tests especially when testing for carbapenem-hydrolyzing Ambler Class D Beta-Lactamases (CHDLs) [14,43].
Limitation
Limitations in this study related to lack of acknowledged fosfomycin breakpoints for bacteria other than E. coli and E. faecalis in the CLSI guidelines. The clinical response of the patients to fosfomycin treatment was also difficult to be reported in present study because not all patients have received fosfomycin treatment and most of patients have already been discharged from hospital. We performed fosfomycin susceptibility screening only to address it as a new alternative agent but clinical data were not sufficient and further studies are needed. Therefore, our recommendations were to provide supportive clinical data in future studies.
Conclusion
The modified CarpaNP test is a rapid test for early detection of carbapenemase producers with excellent sensitivity and specificity for NFGN uropathogens. The mCIM test is a simple and easy to perform test that uses readily available laboratory supplies for recovery of carbapenemase-producing P.aeruginosa but not for Acinetobacter spp. The CarbAcineto NP test provides a rapid and cost-effective solution for carbapenemase detection in Acinetobacter spp., its use will be interesting particularly for ICU patients, for whom MDR Acinetobacter spp. are a common source of severe infections. By using both the mCIM and CarbAcineto NP tests, any microbiology laboratory may have the opportunity to identify one of the most important clinical resistance traits of modern microbiology i.e., carbapenem resistance-mediated mechanisms in clinically significant NFGNB. Fosfomycin is a promising new alternative antimicrobial agent for treatment of MDR uropathogenic NFGNB; however supportive clinical data regarding patient’s response to fosfomycin therapy is a must.