Congenital spine deformities are one of the actual and complex problems in modern orthopaedics and vertebrology. The cause of the vertebral column congenital curvature is anomalies in the development of vertebral bodies during first six weeks of pregnancy [1,2]. Although the incidence of the spine congenital malformations is relatively low (0.5-1 per 1000 newborns) [3,4], but they cause severe deformities in pre-school children, leading to functional impairment of the part of the respiratory and cardiovascular systems. In addition, these congenital spine deformities are one of the leading factors of the disability in the child population.
A sufficient number of studies are devoted to surgical treatment of children with congenital deformity of the spine. In these studies, the postulate has been proved and approved, that operative treatment of these development anomalies of the spinal column is shown at an early age-up to 3 years [5-7]. At the same time, it is known that about 50% of congenital spine deformities progress in the growth process and development of the child and that is why it requires surgical treatment. In view of the foregoing, it is very important to determine and predict: how will the congenital deformity of the spine, revealed in the first year of the child life, behave. One of the most important tasks in this situation is the creation of a complex of diagnostic measures in patients with severe congenital spine deformities, based on the pattern of clinical and radiation studies, and the development of a diagnostic panel based on molecular genetics and biochemical criteria.
According to the literature data, it is known that most often congenital spine deformities arise due to genetic inheritance prerequisites such as various mutations of genes, violations of foetal development for various exogenous or endogenous reasons, disturbances in nutrition of the future mother, lack of vitamins and trace elements, as well as metabolic disorders of the pregnant organism, hormonal disorders in the pregnant woman severe toxicosis, nephropathy and gestosis [8-17].
Modern ideas about the role of teratogenic factors in the congenital malformations genesis are based on the fact that the effects of external influence are most often realised as a result of combined influence of several factors [18]. Both the susceptibility of pregnant women and the agent acting at the appropriate dose at the critical moment of embryogenesis has a damaging effect [19,20].
Congenital scoliosis usually progresses faster than other types of scoliosis (idiopathic scoliosis) and requires surgical treatment [21]. Identification of genetic factors in congenital scoliosis aetiology gives an opportunity to understand the further pathogenesis of the disease, to predict the course of its development and prevent relapses that arise after surgical intervention.
The genetic basis of the aetiology of congenital scoliosis can be determined by mutations in several groups of genes. The genes such as CYP1A1, CYP1A2, GSTT1, GSTM1, NAT2 control the activity of detoxification enzymes that metabolise mutagenic and teratogenic environmental factors to harmless compounds. Mutations in this group of genes can hamper the neutralisation of teratogenic factors and lead to various violations of embryogenesis, including, development of congenital spine anomalies.
In this study, polymorphism in detoxification and DNA repair genes and their relationship to the aetiology and pathogenesis of congenital scoliosis has been determined.
Materials and Methods
Participants
This case control study of detoxification and DNA reparation genes was done on 200 children with congenital deformities of the thoracic and lumbar spine in the age group from 1 year 2 months to 16-year-old who underwent surgical treatment at the Turner scientific research Institute for children’s orthopaedics, St-Petersburg, Russia from September 2015 to May 2018. These children underwent a clinical and radiographic examination. In the structure of the vertebral column’s congenital curvatures, various abnormalities of the development of the vertebrae were encountered: formation disturbances (lateral and posterolateral semi-vertebrae, posterior and lateral sphenoid vertebrae), fusion disorder (asymmetric butterfly vertebrae) segmentation of the vertebrae (blocking the lateral surfaces and front surfaces of the vertebral bodies) and synostosis of the ribs. All patients had a pronounced deformation of the thoracic and lumbar spine with defects in the vertebrae and ribs. These patients were prescribed surgical treatment. Patients with idiopathic scoliosis were excluded from the study.
The control group consisted of 96 healthy children aged from 2 to 16 years without pathology of the spine.
In 8% of the examined patients (n=200), among the accompanying congenital anomalies of other organs and systems were oesophageal atresia, tracheotophageal fistula, renal aplasia, atresia of the anus, congenital crack of the upper lip, congenital anomaly of the tracheobronchial tree, hypoplasia of the lungs, congenital heart defects, etc.
This study was performed according to the Declaration of Helsinki after being approved by the Local Ethics Committee of the Institute. Parents of the all underage participants provided informed consent for their participation in the study.
Genomic DNA Extraction and SNP Selection
Genomic DNA was extracted from whole blood samples using a genomic DNA extraction kit (Interlabservice Ltd., Russia) following the manufacturer’s protocols.
Polymorphism was determined using the PCR method. For preparation of the PCR mixture, Encyclo Plus PCR kit (Eurogen JSC, Russia) and a pair of primers mentioned in [Table/Fig-1] were used.
Structure and properties of oligonucleotide primers.
| Gene and name of primers | Oligonucleotide primers sequence 5’-3’ | Tm, °C | Restriction enzymes; Size of reaction products |
|---|
| CYP1A2°F (-164°A°→°C) | TGAGGCTCCTTTCCAGCTCTCA | 62 | ApaI; 265 bp-A/A, 211+54 bp-C/C, 265+211+54 bp-A/C |
| CYP1A2°R (-164°A°→°C) | AGAAGCTCTGTGGCCGAGAAGG |
| °XRCC1F | CAAGTACAGCCAGGTCCTAG | 63 | AsuC2I; 268 bp-A/A, 177+91 bp-G/G, 268+177+91 bp-G/A |
| °XRCC1 R | CCTTCCCTCATCTGGAGTAC |
| XRCC3 F | GCCTGGTGGTCATCGACTC | 58 | Bsp19I; 136 bp-C/C, 97+39 bp-T/T, 136+97+39 bp-C/T |
| XRCC3 R | ACAGGGCTCTGGAAGGCACTGCTCAGCTCACGCACC |
| NAT2 F | GCCTCAGGTGCCTTGCATTT | 62 | KpnI S1-535 bp, TaqI S2-330+205 bp, BamHI S3-535 bp |
| NAT2 R | CGTGAGGGTAGAGAGGATAT |
| GSTP 114 F | GGGAGCAAGCAGAGGAGAAT | 62 | BspACI; 246+116+58 bp-C/C (wt), 362+246+116+58 bp-C/T, 362+58 bp-T/T |
| GSTP 114 R | CAGGTTGTAGTCAGCGAAGGAG |
| GSTP 105 F | ACCCCAGGGCTCTATGGGAA | 64 | BstMAI; 176bp-A/176+91+85 bp-A/G, 91+85 bp-G/G |
| GSTP 105 R | TGAGGGCACAAGAAGCCCCT |
| GSTT1 F | TTCCTTACTGGTCCTCACATCTC | 62 | 480 bp |
| GSTT1 R | TCACCGGATCATGGCCAGCA |
| GSTM1 F | °GAACTCCCTCAAAAGCTAAAG-C | 62 | 215 bp |
| GSTM1 R | °GTTGGGCTCAAATATACGGTGG |
| °CF94 F | °GTTTTCCTGGATTATGCCTG | 62 | 97 bp |
| °CF94 R | °GTTGGCATGCTTTGATGACG |
PCR for the genes CYP1A2 (-164 A→C), XRCC1, XRCC3, GSTP 114, GSTP 105, NAT2 was performed under the following temperature conditions: 3 minutes-denaturation at 95°C, 35 amplification cycles at 95°C for 30 seconds, Tm-30 seconds, 72°C-30 seconds. PCR was terminated with final extension at 72°C for 7 minutes, followed by cooling to 4°C (Tm see in the [Table/Fig-1]).
To determine the nucleotide substitutions, the Restriction Fragment Length Polymorphism (RFLP) was used. Restriction of the amplified DNA fragments was performed using restriction endonucleases (SibEnzyme Ltd., Russia) according to the manufacturer’s instructions. The frequency of the null allele of the GSTT1 and GSTM1 gene was examined by PCR. A 25 μL amplification mix comprised: 0.25 μl of 100 pmol of each primer, 12.5 μL of 2x PCR buffer, 0.5 μL of dNTP, 0.25 μL of Taq polymerase (Encyclo Plus PCR kit), 1 μL of genomic DNA, H2O to 25 μL.
The results of PCR and RFLP were analysed by DNA gel electrophoresis in 9% polyacrylamide gel followed by coloring in SYBR Green I (Biotech Industry Ltd., Russia) and visualisation of fragments in the UV light.
Statistical Analysis
Results were statistically processed using the software package Statistica 6.0. Group comparisons with respect to categorical variables were performed using chi-square tests or, alternatively, Fisher’s-exact test when expected cell counts were less than 5. A p-value less than 0.05 were considered significant.
Results
The polymorphisms of the genes CYP1A2, NAT2, GSTM, GSTT, GSTP, XRCC1, XRCC3 and their frequency distribution among patients with CSD were investigated [Table/Fig-2].
Allelic variants of the studied genes. Allelic variants of the studied genes.
| Gene | Polymorphism | Genotypes |
|---|
| CYP1A2 | 164A→C | A/A, A/C, C/C |
| GSTM1 | +/0 | +/00 |
| GSTT1 | +/0 | +/00 |
| GSTP1 | Ile105Val | A/A, A/G, G/G |
| GSTP1 | Ala(C)114Val(T) | C/C, C/T, T/T |
| NAT2 | Leu161Leu | *5 |
| NAT2 | Arg197Gln | *6 |
| XRCC1 | Arg399Gln | G/G, G/A, A/A |
| XRCC3 | Thr241Met | C/C, C/T, T/T |
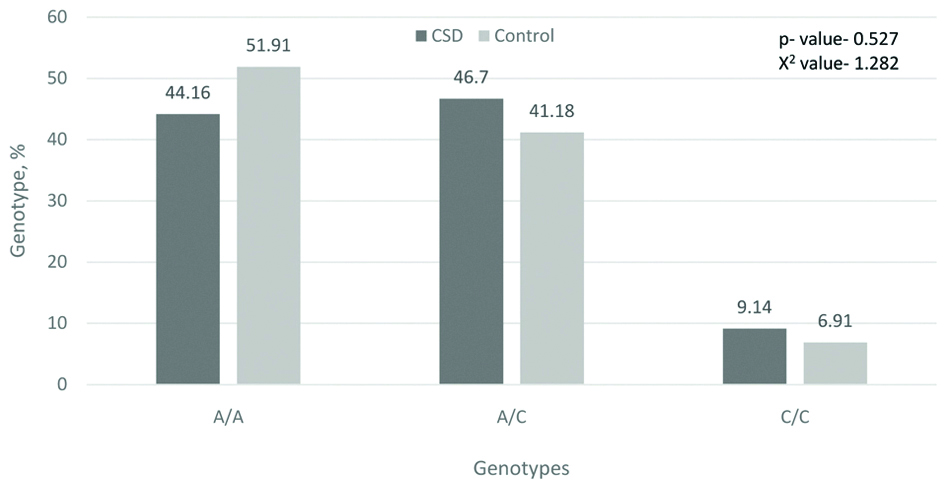
From the literature data it is known that the presence of the C-allele characterises the slow metabolism of xenobiotics during induction with caffeine, omeprazole and smoking [22]. In our study, 55.8% of patients with CSD had an allele of a slow metaboliser, while in the control group this value was 48.09%[Table/Fig-3].
Frequency of genotypes by the gene CYP1A2 164A→C, %.
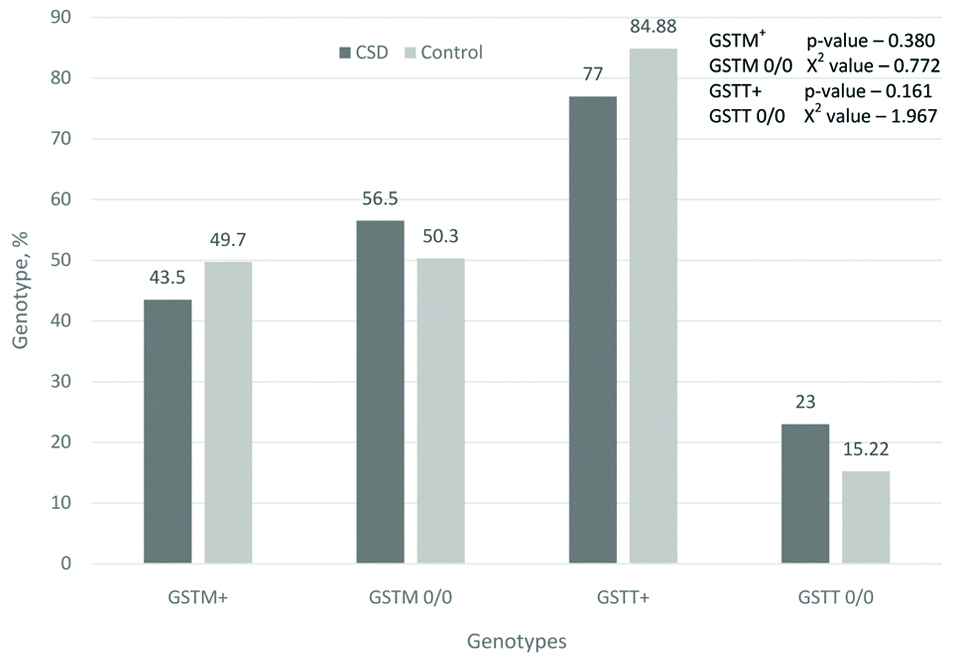
Homozygotes for the normal “+” allele were determined by the presence of an amplification product of 480 bp for GSTT1 and a fragment of 215 bp for GSTM1 on the electrophoregram. The absence of a corresponding fragment indicated the homozygosity of the individual for the gene deletion. Deletion of glutathione-S-transferase genes results in the absence of these enzymes in the body and, accordingly, the possible accumulation of xenobiotics, which are teratogenic factors that contribute to the emergence of CSD. In the group of examined patients, 79.5% had deletions of either GSTM1 or GSTT1 genes, and amongst them 13.5% of cases had deletion of both genes. This can be an important factor in the aetiology of the CSD. In the control group, the deletion of one or another gene of this group was 65.77%, and both genes was 9.47%, which is much less [Table/Fig-4].
Frequency of genotypes by genes GSTM1 and GSTT1, %.
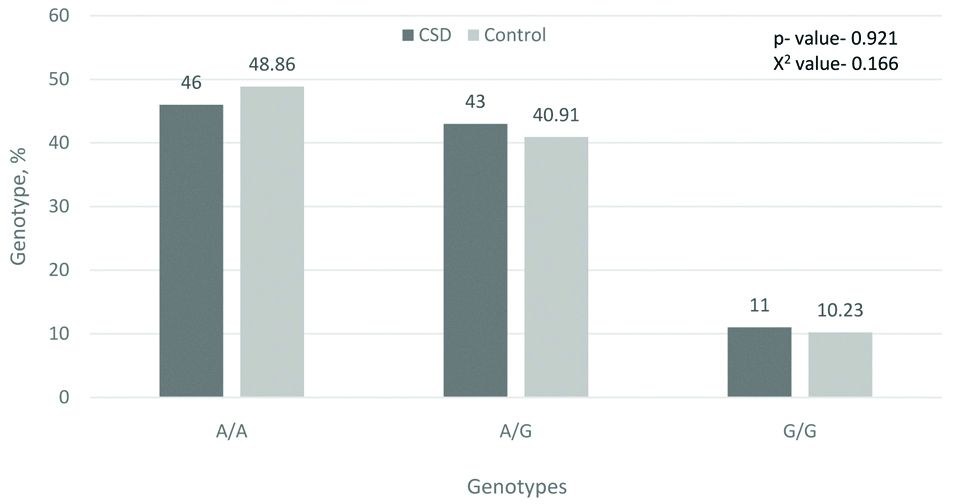
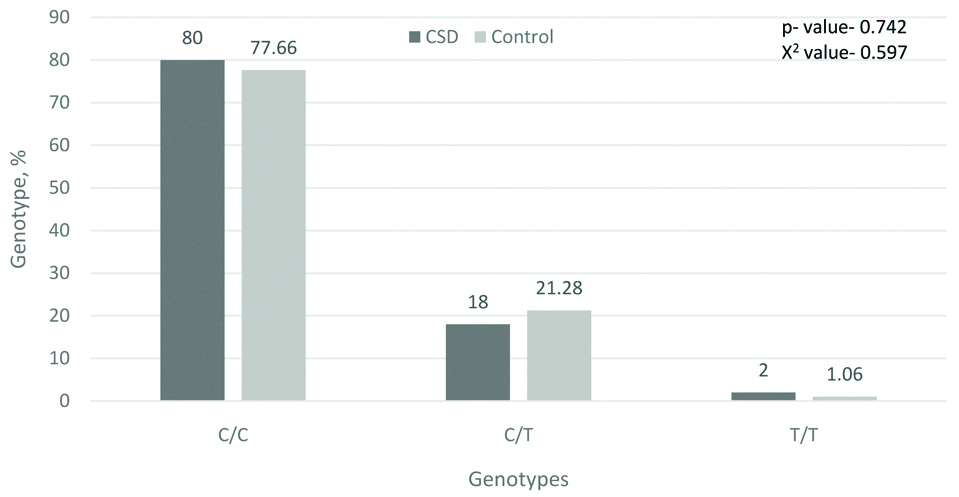
As mentioned above, polymorphisms of 105Val and 114Val of the GSTP1 gene decrease enzyme activity and increase sensitivity to the action of mutagens and carcinogens. Our study did not reveal significant differences in the distribution of GSTP1 genotypes in patients with CSD and in the control group, but 8.5% of patients with CSD had a co-carrier homozygous for the minor allele with the null allele of the GSTM1 gene, which was not seen in the control group. This can also serve as one of the predisposing factors to the emergence of CSD [Table/Fig-5,6].
Frequency of genotypes by gene GSTP1 (Ile105Val), %.
Frequency of genotypes by gene GSTP1 (C114T), %.
In the study of the NAT2 gene, we found an increase of 35.46% (9.13% versus 6.74%) in the frequency of the homozygous genotype of the slow acetylator of the * 6/6 allele (G590A) in patients with CSD compared with the control group.
When studying the alleles of polymorphism of Arg399Gln of the XRCC1 gene, no significant differences from the control group were found, however, the percentage of the mutant allele in the homozygous state in patients with CSD was higher.
For the XRCC3 gene, the difference in the distribution of allelic variants of the Thr241Met polymorphism between patients with CSD and the control group was more significant. Homozygotes in the major normal allele among patients with CSD were 53.64% (in the control group-78.13%). Heterozygous carriage was detected among 33.66% of patients with CSD (in the control group-13, 54%). The content of the homozygous mutant allele among the patients with CSD was 12.7% (in the control group-8.33%).
Discussion
The emergence of congenital malformations, like any multifactorial pathology, is associated both with the impact of unfavorable teratogenic factors of the environment during pregnancy (hypoxia, a number of drugs, occupational hazards, alcohol use, hyperthermia, insulin-dependent diabetes mellitus and gestational diabetes) [8-14], and genetic factors (chromosomal aberrations, gene polymorphisms associated with hereditary predisposition, de novo mutations, epigenetic changes) [15-17,23]. Each of these factors alone or a combination of them can lead to a violation of embryogenesis and anomalies in the development of the vertebrae. Recently, studies have been done on the analysis of molecular-genetic markers accompanying congenital spine deformities. In these studies, possible factors of the aetiology and pathogenesis of congenital spine deformations, such as mutations in PAX1, DLL3, SLC35A3, WNT3A, TBX6, and T (Brachyury) genes are considered in sufficient detail [4,24,25]. The role of heredity in the onset of congenital scoliosis is also evidenced by the fact that this disorder occurs in monozygotic and dizygotic twins in conjunction with other congenital malformations [26,27]. Possible mechanisms in this process are epigenetic factors characterised by abnormal methylation [28].
The investigation of detoxification and reparation genes was previously carried out for various hereditary and exogenous disorder. In particular, such studies were most often carried out in oncology patients with different aetiology of the tumour and a relationship with predisposition to the disorder was shown [29-31]. In fact, the modification in the activity of detoxification and reparation genes leads, first, to the fact that the body accumulates more teratogenic factors that can toxically affect metabolism, causing damage to organs and tissues. It can be sensitive to the body of a pregnant woman and the foetus that she bears. There are also publications on this subject that discuss the correlation between the activity of detoxification and repair genes with the occurrence of congenital malformations [32-34]. Mutations in the genes of DNA repair can directly lead to chromosomal and gene anomalies [35,36]. When analysing the most significant genome defects (chromosome aberrations), it was found that microdeletions in the chromosome region of 17q21.31 [37], 16p11.2 [38] lead to congenital deformities and injuries of the spine. In these cases, it is interesting to note that in the deletion region 16p11.2 there is TBX6 gene, which is a member of the T-box gene family - transcription factors regulating, in particular, somitogenesis and spine ontogenesis [39]. Therefore, the study of the aetiology of congenital spine deformities inevitably leads to the need to investigate the genes of detoxification and reparation. Due to time limitation and labor-extensive nature all the genes of these systems, could not be analysed. The results thus obtained indicate a correlation between mutational changes in these genes and the occurrence of congenital spine deformities. In particular, the significant changes in the genes CYP1A2, GSTM1, GSTT1, NAT2, XRCC3 revealed that these genes, and hence, the DNA detoxification and repair system itself, are involved in the process of protecting somitogenesis from teratogenic disorders. If this protection system is weak and has gaps, congenital spine deformities can occur with a high degree of probability.
This study confirms the fact that the mutations in the DNA repair genes can lead to chromosomal and gene anomalies. This data is supported by the results of literature sources [3,40,41], which emphasize that the majority of patients with congenital deformities of the spine have a combination of defects in the development of other internal organs and systems, which is associated with chromosomal aberrations in the linkage group to other genes.
Further studies will be carried out to identify the most significant polymorphisms of the detoxification and DNA repair genes that most significantly affect the violation of spine embryogenesis, causing the appearance of disorders such as impaired formation, fusion and segmentation of the vertebrae which leads to severe progressive spine deformations.
Limitation
Because of the limited time, we study the polymorphism of all the genes of the detoxification and reparation systems could not be studied.
Conclusion
The study of patients with congenital spine deformities made it possible to identify a part of the unfavorable genetic load contributing to the emergence and progression of this severe pathology. It was determined that children with multiple and combined defects of the spine development have more mutations in detoxification and DNA repair genes, while the simultaneous carriage of several mutant alleles in CSD patients is more than twice that in the control group. The obtained results allow to some extent to assume the nature of the course of the congenital spine deformities in patients at an early age. However, the final evaluation and identification of molecular genetic criteria for the progression of the congenital spine deformities in children requires further study.