Paediatric ALL is the most prevalent childhood cancer, with around 2 per 100,000 newly diagnosed cases per year all over the world [1]. It is a cancer of the bone marrow and blood cells, where cancerous cell replicates uncontrollably and become resistant to cell death. Acute leukaemia progresses rapidly and may result in death within two or three months if left untreated [2]. ALL is a prototype of circulatory cancer that can be treated with combination chemotherapy alone [3]. The vast majority of the literature is focused on anti-cancer drug 6-MP metabolism used for ALL treatment, because of its relatively high prevalence and excellent survival rate in children [4]. Thiopurine drugs are purine analogue which is metabolised inside the human body to form TGNs metabolites, which have cytotoxic and immunosuppressive properties [5].
TPMT is a cytosolic enzyme encoded by TPMT gene that catalyses the S-methylation of thiopurine. It is responsible for the inactivation of 6-MP, and the formation of non-cytotoxic methylmercaptopurine (6-MMP) [6]. The patients with low TPMT enzyme activity have low 6-MMP formation and much higher 6-TGNs concentration that leading to liver and bone marrow toxicity [7]. Patients treating with standard doses of 6-MP can develop life threatening bone marrow toxicity as a result of excess TGN metabolites production [8].
Materials and Methods
This cross-sectional case-control study included 81 patients being treated for Acute Lymphoblastic Leukaemia during the maintenance phase. The sample constituted 59 boys and 22 girls of the age group between 1.83-16.25 years, all of them were receiving 6-mercaptopurine as daily treatment for ALL as per National Randomised Trial of UKALL 2011 MRC protocol (United Kingdom Acute Lymphoblastic Leukaemia-Medical Research Council) for treatment of Acute Lymphoblastic Leukaemia in Children and Young Adults. They were treated and followed by the oncology unit and oncology outpatient clinic in Children Welfare Teaching Hospital, Medical City, Baghdad during the period March 2017 to July 2017. A verbal informed consent was obtained from all the patients’ guardians after being informed about the utility of the study.
Genotyping
Two mL of peripheral whole blood in EDTA tube was taken from each patient for the genotyping analysis and were stored at -20°C until further use.
DNA Extraction
The extraction of the genomic DNA from the whole blood was done using ExiPrep™ Plus Blood Genomic DNA Kit (K-4211) provided by Bioneer, Korea. The DNA was isolated as per the manufacturer’s instructions. The samples were analysed by agarose gel electrophoresis to ensure their suitability for PCR analysis and stored at -20 °C until use.
Polymerase Chain Reaction (PCR)
The PCR reaction was carried out usingof AccuPower PCR PreMix (Bioneer, Korea) for the detection of TPMT*1 gene, and AccuPower Gold Multiplex PCR Premix (Bioneer, Korea) for the detection of TPMT*3A, TPMT*3B and TPMT*3C genes. The PCR PreMix consisted of DNA polymerase, dNTPs (dATP, dCTP, dGTP, dTTP), reaction buffer with a MgCl2 solution, stabiliser and tracking dye and in addition to these, the Multiplex PCR PreMix consisted of pyrophosphatase with pyrophosphate. The reaction mixture contained 10 μL of premix, 5 μL DNA and 1 μL of each primer (forward and reverse) and 8 μL of distilled water to obtain the final volume of 25 μL.
PCR amplification consisted of initial-denaturation step at 95°C for five minutes followed by 35 cycles of denaturation at 95°C for one minute, annealing at the specified annealing temperature for two minutes, followed by extension at 72°C for one minute and the final extension step at 72°C for five minutes. The PCR amplicons were separated on 2% agarose gel through electrophoresis. The bands were visualised with an Ultraviolet (UV) transilluminator at 365 nm (Cleaver Scientific, UK).
Detection of Wild-Type (TPMT*1)
The TPMT*1 allele was analysed by allele-specific PCR, using sequence-specific primers:
P2FW 5’GTATGA TTT TAT GCA GGT TTG 3’ (forward), and
P2R 5’TAA ATA GGA ACC ATC GGA CAC 3’ (reverse)
DNA fragment was amplified with P2FW and P2R primers by PCR assay with an annealing temperature of 53°C for two minutes.
Detection of TPMT*3A, TPMT*3B and TPMT*3C Mutations
TPMT mutant alleles were detected by allele-specific multiplex-PCR with specific primers synthesised by Bioneer, Korea [Table/Fig-1]. Genotype analysis for G460 A point mutation at exon 7 was carried out by PCR assay with an annealing temperature of 59°C for two minutes and was amplified using primers sequence of 460F and 460R [Table/Fig-1]. The 338 base pairs fragment yield from the PCR amplification were detected by electrophoresis technique in 2% agarose gel. Genotype analysis for A719G point mutation at exon 10 was carried out by PCR assay with an annealing temperature of 56°C for two minutes using primers with a sequence of 719F and 719R [Table/Fig-1]. The 273 base pairs fragment yield from the PCR amplifications were detected by electrophoresis technique with the use of 2% agarose gel. Genotype analysis for A719G point mutation at exon 10 for TPMT*3C allele was carried out by PCR assay with an annealing temperature of 56°C for two minutes using primers with a sequence of 719F and 719R [Table/Fig-1]. The 273 base pairs fragment yield from the PCR amplifications were detected by electrophoresis technique with the use of 2% agarose gel.
Primers used for TPMT*3A (SNP in Exon 7 and 10), TPMT*3B(SNP in Exon 7), and TPMT*3C (SNP in Exon 10) mutation and their products’ length. (bp: base pair, F: Forward, R: Reverse, and SNP: Single nucleotide polymorphism).
| Exon | Primer’s Name | Sequence of Primer | Product Length (bp) | Annealing Temperature (C°) |
|---|
| 7 | 460F | 5’-GGGACGCTGCTCATCTTCT-3’ | 338 | 59 |
| 460R | 5’-GCCTTACACCCAGGTCTCTG-3’ | 338 | 59 |
| 10 | 719F | 5’-AAGTGTTGGGATTACAGGTG-3’ | 273 | 56 |
| 719R | 5’-TCCTCAAAAACATGTCAGTGTG-3’ | 273 | 56 |
TPMT Phenotype
Two mL of peripheral blood was obtained from all patients’ in-plane tube and was clotted at room temperature. It was then centrifuged at 3600 rpm for 10 minutes. The obtained sera were frozen at -20°C in Eppendorf safe-lock tubes until used. This assay was performed using the TPMT ELISA Kit provided by MyBioSource, USA, with Sandwich-ELISA plates that have been pre-coated with an antibody specific to the TPMT enzyme.
Statistical Analysis
Statistical analyses were done by using the SPSS version 22.0 for Windows, (SPSS Inc., Chicago, IL) [15]. Data were evaluated using a Two-Sample Student’s t-test to determine the difference in allele frequencies proportion in TPMT polymorphisms. Pearson correlation analysis was done to determine the correlation between TPMT enzyme activity and genotype. The level of significance, p-value <0.05 was considered statistically significant.
Results
In this study, authors detected three most common inactive TPMT alleles (TPMT*3A, TPMT*3B and TPMT*3C). A total of 81 ALL paediatric patients that receiving 6-MP drug during their maintenance phase of treatment protocol were enrolled in the study. The age of children ranged between 1.83-16.25 years (starting from 1 year and 10 months to 16 years and 3 months) of age, with age mean 7.08±3.12 years were studied. The incidence of ALL in Iraqi children is high and it is much higher in boys than in girls. ALL children patients were 59 boys and 22 girls [Table/Fig-2].
Genotype frequencies of TPMT variants in a sample of 81 Iraqi paediatric ALL patients (SNP: Single nucleotide polymorphism; rs: recessive spotting).
| Allele | SNP Position | rs | Amino acid Substitution | Gender | Allele No. | Frequency % |
|---|
| M | F |
|---|
| TPMT*1 | Wild-type | | | 37 | 14 | 51 | 62.96 |
| TPMT*3A | G460AA719G | 18004601142345 | Ala 154 ThrTyr 240 Cyc | 18 | 6 | 24 | 29.62 |
| TPMT*3B | G460A | 1800460 | Ala 154 Thr | 0 | 0 | 0 | 0.00 |
| TPMT*3C | A719G | 1142345 | Tyr 240 Cyc | 4 | 2 | 6 | 7.4 |
| Total | 59 | 22 | 81 | |
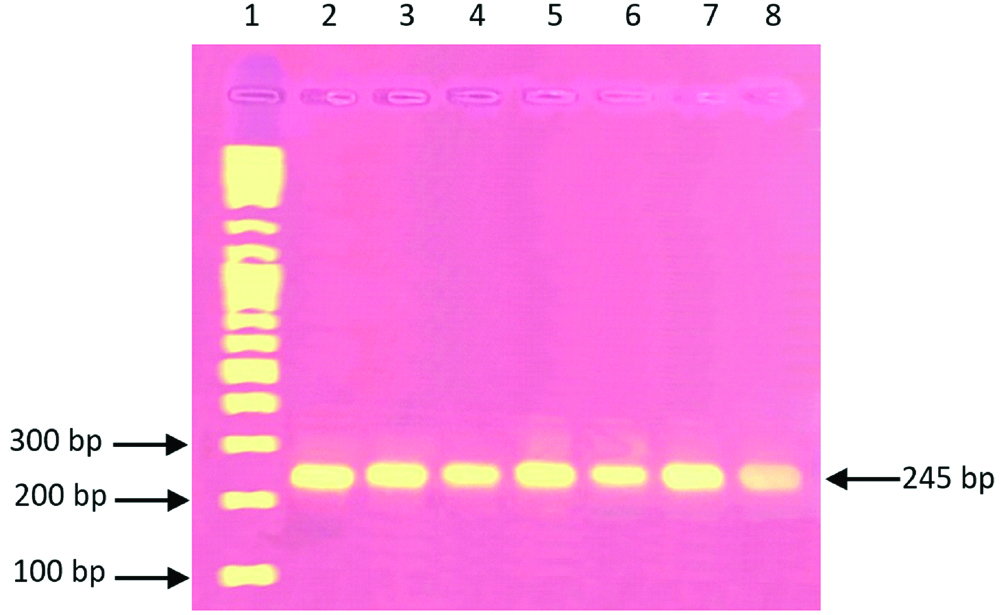
A total of 51 children with allele frequencies of (62.96%) were homozygous for the wild-type allele TPMT*1 which mean that they did not carry any of the three detected mutant alleles. When wild-type was present the PCR reaction yielded 245 base pairs band [Table/Fig-3]. The band with 245 bp that corresponding to TPMT*1 allele was presented in all the samples revealing that all the tested samples were either wild-type (have two normal alleles) or heterozygote (have one normal allele and one mutant allele) for TPMT mutant gene. Thirty children with an allelic frequency of 37.03% were heterozygous for one of the two mutant alleles (TPMT*3A or TPMT*3C), while TPMT*3B allele was not detected in this studied children’ population.
Electrophoresis patterns for TPMT alleles analysed by allele-specific PCR. Lane 1 corresponding to 100 bp DNA ladder on a 2% agarose gel; lanes 2-8 show 245 bp PCR product using primers P2R and P2FW corresponding to TPMT*1 (wild-type) allele.
Genotype analysis for TPMT*3A allele was identified by the presence of two bands (273 and 338 bp) referred to the presence of G460A and A719G SNPs together in the same sample [Table/Fig-1]. The presence of a band at 273 bp was corresponding to TPMT*3C allele [Table/Fig-4].
Electrophoresis patterns corresponding to migration of different alleles of TPMT analysed by Allele-Specific Multiplex PCR. Lane 1 showed a 100 bp DNA ladder on a 2% agarose gel. Lane 4 shows two fragments of 338 and 273 bp corresponds to TPMT*3A polymorphism. Lanes 6 and 7 show a single fragment of 273 bp corresponds to TPMT*3C polymorphism. Lane 2, 3, 5 and 8 show no PCR fragments.
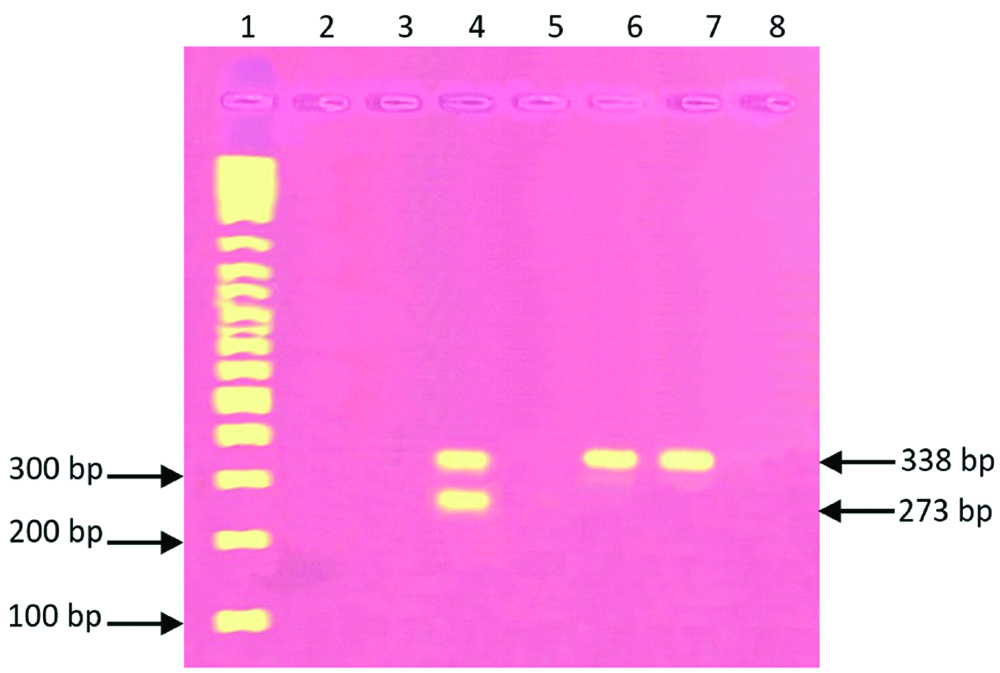
Twenty-four children had TPMT*3A mutant allele leading to allele frequencies of 29.62%, and six children had TPMT*3C mutant allele leading to allele frequencies of 7.4% [Table/Fig-2].
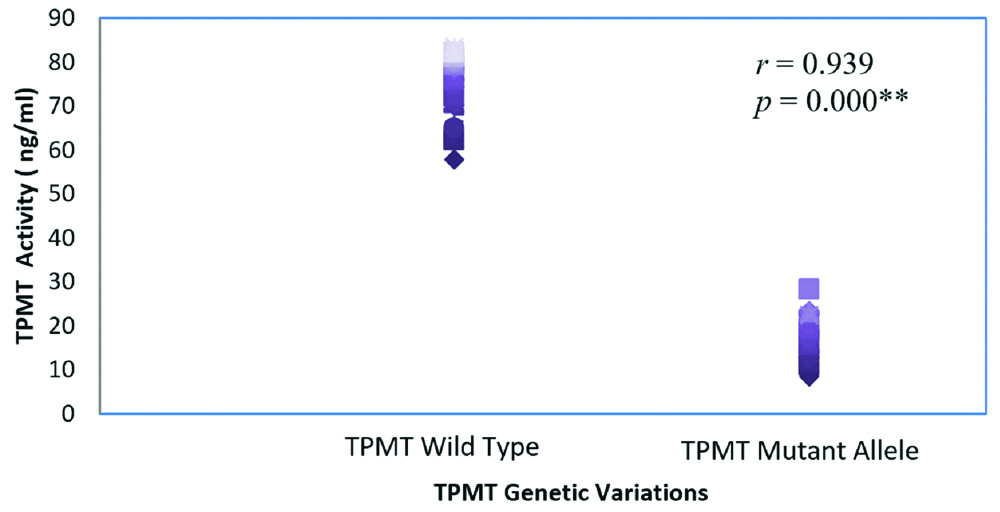
In total, 51 ALL children patients the TPMT*1 activity ranged from 57.86 to 83.41 ng/mL with mean±SD of 76.27±6.53 for the wild-type TPMT allele in comparison to activity of mutant allele of TPMT gene which ranged from 8.41-28.322 ng/mL with mean±SD of 24.13±17.63. The difference in mean of TPMT activity between ALL patients carrying the wild-type gene and ALL patients carrying the mutant gene was statistically highly significant (p-value ≤0.001) [Table/Fig-5].
TPMT activity (ng/mL) in Iraqi children according to TPMT genetic variation.
| TPMT allele | n | TPMT activity range (ng/mL) | Mean±SD | p-value |
|---|
| TPMT*1 | 51 | 57.86-83.41 | 76.27±6.53 | ≤0.001** |
| TPMT*3A and TPMT*3C | 30 | 8.41-28.322 | 24.13±17.63 |
| TPMT*3A | 24 | 8.407-28.322 | 16.83±4.69 |
| TPMT*3C | 6 | 15.571-23.063 | 23.96±9.74 |
The results of genotyping and phenotyping methods showed a highly significant positive correlation between TPMT low activity and presence of genetic mutation across the TPMT gene (p≤0.001). The relationship was very strong with a positive value of r>0.5 (0.939), [Table/Fig-6].
The association between TPMT enzyme phenotype and genotype; TPMT enzyme activity in ALL paediatric patients with mutant allele TPMT*3A and TPMT*3C compared to TPMT wild-type allele.
Discussion
The most common cause of morbidity and mortality among children with ALL in Iraq is infection [16,17]. Increased risk of infections has been associated with neutropenia as part of myelosuppression in ALL patients receiving 6-MP during chemotherapy. Many patients with ALL during maintenance were scheduled to reduce their assigned treatment dosage because of persistent neutropenia. Treating suspicious patients with adjusted dosage of 6-MP can be of great clinical importance to avoid unnecessary toxicity.
TPMT activity is known to exhibit genetic polymorphism in most populations studied to date, and the genetic basis for this inherited trait has been explained in most ethnic groups and races worldwide. Among various races, the Arab race is considered one of the distinct races [18]. The present study is the first to elucidate the genetic basis of TPMT inherited trait in the Iraqi paediatric ALL population using the genotype and phenotype analysis.
The best model for incorporating pharmacogenetics with the clinical practice is TPMT polymorphism. TPMT activity is inherited as an autosomal co-dominant trait with large inherited variations in human tissue [19].
Variant alleles of the TPMT gene have been associated with low TPMT activity, therefore for the safety and efficacy of ALL treatment; the TPMT genotype and phenotype are considered a useful tool for optimizing 6-mercaptopurine drug. In the present study, ALL patients with TPMT mutant allele have low TPMT activity as compared with those having wild-type allele, as a reason for low enzyme level in those patients. Accordingly, phenotypic analysis may be useful and could be performed concomitantly with genotyping tests [20].
Three variant TPMT alleles; TPMT*3A, TPMT*3B, and TPMT*3C, were genotyped and their frequencies were determined. The TPMT*3A and TPMT*3C alleles were detected in 30 patients out of 81 ALL paediatric patients during the maintenance phase under 6-MP therapy, the total frequencies of TPMT mutant alleles were 37.03%. However, the present study has shown that none of the ALL children; included in the present study, carried the TPMT*3B mutant allele. TPMT*3B is a rare allele that is usually absent in most populations [18,20-22].
In the current study, TPMT*3A allele had shown a higher frequency than TPMT*3C allele. These results are comparable to the Caucasian population, where frequency of TPMT*3A was 60-89% of total deficiency alleles, while TPMT*3C mutant alleles are only 5-15% [21].
For the Jordanian population, only the TPMT*3A and TPMT*3C mutant alleles were detected, with an allele frequency of 0.59% and 0.30% respectively, this is similar to present finding [23]. In Palestine, only TPMT*3A allele was detected with a frequency of 0.89% [24]. This finding comes in line with the results of all studies conducted in Asian countries and the majority of studies conducted at Middle-Eastern countries [22,24-27]. A study of the mutant allele in the Libyan population revealed the presence of TPMT*3A and TPMT*3C and the allele frequencies were 0.61% and 1.02% respectively [10].
A study of the mutant allele in the Egyptian population found that TPMT*3A was the most frequent allele of 56%, also the TPMT*3C and TPMT*3B mutant alleles were found with a frequencies of 16% and 8% respectively [25]. In Turkish children with acute lymphoblastic leukaemia, TPMT*3A and TPMT*3C were the only TPMT genetic polymorphism found with allele frequencies of 3.4% and 0.9% respectively, and no cases were identified with TPMT*3B or TPMT*2 variant alleles [26], this study comes in line with the present results. A study on Iranian population found that TPMT*2 is the most common mutant allele with a frequency of 3.94%, their study, also, indicated the presence of mutant alleles TPMT*3A and TPMT*3C with allele frequencies of 0.79% and 1.57% respectively, Azad M et al., [27].
The current study showed a higher frequency of defective TPMT*3A and TPMT*3C alleles in comparison with other studies mentioned, this is largely due to the sample chosen by the investigator and the supervising physician in which 42 patients (out of the total 81 patients) were labelled as intolerant during the course of the disease either because of persistent neutropenia or Jaundice (with negative other investigations) and dosing of the oral chemotherapy (6MP) was adjusted frequently to render them able to continuously receive their assigned treatment but with a lower dosing. Only 12 patients out of the 42 intolerant patients showed were homozygote for wild-type (TPMT*1) allele and no defective alleles were recognised in their phenotypic and genotypic analysis. The absence of predominant types of TPMT enzyme mutations and the presence of myelosuppression in these three patients may result from the existence of other mutant alleles of TPMT or mutation in other enzymes involving in 6-MP metabolism or other non-genetic factors [24]. The other 39 tolerant patients, were chemotherapy was given in their standard dosing, showed only two patients with TPMT*3A defective allele while all other cases were homozygous for the wild-type.
Limitation
The TPMT activity assay has some limitations. It cannot be applied on subject that recently has received a blood transfusion. Moreover, the interactions between certain drugs (e.g., methotrexate), alcohol, and food (e.g., milk) can lead to false results. Furthermore, physiological factors such as gender, age and red blood cells lifespan can impact the TPMT activity and affects the results. Further studies are needed for healthy adult subjects and leukaemia patients to evaluate the TPMT activity and detect the most common TPMT polymorphism in Iraqi population.
Conclusion
TPMT*3A and TPMT*3C were the only deficiency alleles detected in the Iraqi paediatric ALL patients with low TPMT activity detected in their serum. Therefore, the TPMT genotype together with phenotype can provide an important molecular biomarker for predicting the response of the high-risk group to the anti-cancer therapies and minimise the toxic effects of thiopurine drugs. The current study demonstrates that one of the myelosuppression causes of the Iraqi paediatric ALL patients treated with 6-MP was a low TPMT activity that related to the TPMT genetic polymorphisms exists in these patients.