Introduction
Dengue fever is one of the most important arboviral infections all over the world. Globally, 50% of the human population is at risk of dengue virus infection. Circulation of more than one serotype and virulent genotypes are responsible for major outbreak with high mortality in any geographical region.
Aim
To identify the Dengue virus serotypes and genotypes circulating in Chennai, Tamil Nadu, India.
Materials and Methods
Dengue serum samples were subjected to Reverse Transcriptase-PCR and representative samples were sequenced, subsequently; Maximum-Likelihood phylogenetic tree was constructed by MEGA 7 software.
Results
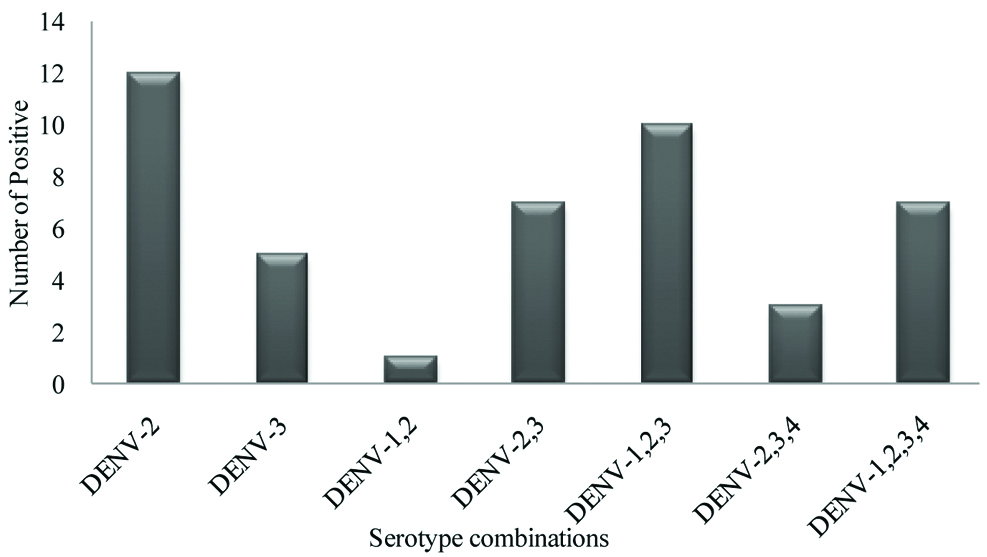
Out of 140 samples tested, 45 were positive for different serotypes of dengue virus. Among the 45 PCR positives, 12 (26.7%) and 5 (11.1%) samples were positive for Dengue virus-2 and Dengue virus-3 respectively. The remaining 28 (62.2%) samples showed more than one serotype infection. The following combinations of multiple infections were observed in the present study: one sample (2.2%) had mixed infection of Dengue virus-1, 2; seven samples (15.5%) had mixed infection of Dengue virus-2, 3; 10 samples (22.2%) had mixed infection of Dengue virus-1, 2, 3; three samples (6.7%) had mixed infection of Dengue virus-2, 3, 4 and seven samples (15.6%) had mixed infection of all four serotypes. The predominant serotypes were Dengue virus-2 and 3 when multiple serotypes were concerned. Genotyping analysis revealed that Dengue virus-1 sequence belonged to Genotype-I, Dengue virus-2 belonged to Genotype-IV, Dengue virus-3 belonged to Genotype III.
Conclusion
In the present study, authors conclude that the circulation of multiple serotypes with a high number of concurrent infections, re-emergence and continuous circulation of genotypes in our region. The burden of dengue viral disease in India is massively under-reported. Hence detailed continuous molecular surveillance studies and stringent control measures are urgently warranted to save lives from dengue-related mortalities.
Introduction
Dengue virus is an arthropod-borne, enveloped, single-stranded, positive-sense RNA virus of the Flaviviridae family which causes emerging and re-emerging infections throughout the world [1]. Dengue virus consists of four antigenically different serotypes (DENV-1–4) and the fifth serotype Dengue virus-5 was announced in the year 2013 in Malaysia [2]. Within the serotypes, several genotypes have been reported based on their genetic diversity and geographical distribution [3].
Globally, there are 128 countries at risk of infection with Dengue virus [4] and an estimated 390 million infections occur every year, of which 96 million manifested as a clinically severe disease [5]. Dengue infection is endemic in India and causing more frequent outbreaks. A total of 596467 cases and 1542 deaths were reported during the period of 2010-2017. Delhi, Haryana, Gujarat, Karnataka, Kerala, Madhya Pradesh, Maharashtra, Orissa, Punjab, Rajasthan, Tamil Nadu, Uttar Pradesh and West Bengal are the severely affected states [6].
Hyperendemic circulation of multiple serotypes is one of the most significant factor responsible for severe dengue disease [7]. Circulation of all four serotypes was first reported during the 1968 outbreak in Vellore. Dengue virus-2 was the predominant serotype from the 1970s to 2000 and it was responsible for 1993 and 1996 outbreaks. Dengue virus-3 was the major serotype responsible for 2003 and 2006 outbreak in Delhi. Dengue virus-1 and 4 have not been associated with any major outbreak in the country [8]. Very few studies were reported on the serotypes circulating in Tamil Nadu, India [9,10], whereas it is rarely reported in Chennai. Considering the incidence, severity and limited treatment options, periodic Dengue virus studies are important. Capsid pre-membrane junction (C-prM) is widely used for serotyping due to the presence of epidemiologically important sequence [11]. Hence, the present study aimed to identify the Dengue virus serotypes and genotypes circulating in Chennai, Tamil Nadu, India using C-prM gene PCR.
Materials and Methods
Study Design
A cross-sectional study was designed to elucidate the serotypes and genotypes circulating in Chennai, Tamil Nadu, India during the period of 2013-2015. The study protocols were approved by the Institutional Human Ethics Committee of DR. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras (IHEC NO: /UMIHEC/05-2013-I).
Collection of Dengue Positive Serum Samples
A total of 140 NS1 positive serum samples from patients who visited Hitech Diagnostic Center and ESI Hospital, Chennai, Tamil Nadu, India with symptoms of dengue fever were included in the present study. The major exclusion criteria adapted was IgM alone positive samples.
Viral RNA Extraction and cDNA Synthesis
Viral RNA was extracted using the QIAamp Viral RNA extraction kit (Qiagen, Germany) according to the manufacturer’s protocol. RNA was quantified by NanoDrop 2000 (Thermo Scientific Inc.) and stored at -80°C until use. Viral RNA was converted to cDNA by SuperScript® III First-Strand cDNA Synthesis kit (Cat No: 18080-400) as described by the manufacturer.
Nested PCR for Serotyping
Nested PCR was performed for C-prM junction of dengue virus. Two rounds of amplification were carried out according to Lanciotti RS et al., [12]. Twenty microlitre reaction was standardised as follows: 2 μL of cDNA with PCR mixture containing each 2 μL (10 pmol) of forward D1 and reverse D2 primers, 2U of Taq DNA polymerase, 2 μL of 10X buffer (Tris-HCl pH 9.0), PCR enhancers, (NH4)2SO4 and 20 mM MgCl2), 10 mM dNTP mix and 11.4 μL (GenetBio Korea) of nuclease-free water. The reaction was amplified in Eppendorf Master Cycler gradient (Germany). Second round PCR was performed for first-round PCR products with dengue serotype-specific primers TS1F, TS1R for DENV-1, TS2F, TS2R for DENV-2, TS3F, TS3R for DENV-3 and TS4F, TS4R for DENV-4 [13]. Primer sequences and cycling conditions were given in [Table/Fig-1] [12,13].
Primers and cycling conditions used for Nested PCR [12,13].
| Primer | 5’-3’ sequence | Cycling condition | Product size |
|---|
| D1-D | TCAATATGCTGAAACGCGWGAGAAACCG | Initial denaturation-94°c for 3 minutes Final extension-72°c for 5 minute Final extension-72°c for 5 minute | 511 bp |
| D2-D | TTGCACCARCARTCWATGTCTTCWGGYTC |
| TS1-F | AGGACCCATGAAATTGGTGA | 411 bp |
| TS1-R | ACGTCATCTGGTTCCGTCTC |
| TS2-F | AGAGAAACCGCGTGTCAACT | 403 bp |
| TS2-R | ATGGCCATGAGGGTACACAT |
| TS3-F | ACCGTGTGTCAACTGGATCA | 453 bp |
| TS3-R | CAGTAATGAGGGGGCATTTG |
| TS4-F | CCTCAAGGGTTGGTGAAGAG | 401 bp |
| TS4-R | CCTCACACATTTCACCCAAGT |
Nucleotide Sequencing and GenBank Submission
Sequencing was outsourced to Amnion Biosciences Bangalore, India, to analyse the gel eluted PCR product in ABI 3730 Genetic analyser using BigDye® Terminator v3.1 Cycle Sequencing Kit. The homology search was performed for the sequences using NCBI BLAST Database. Sequences were submitted to GenBank and accession numbers were obtained.
Phylogenetic Analysis of Dengue Virus Sequences
Phylogenetic analysis was performed by MEGA 7. The Maximum-Likelihood phylogenetic tree was constructed for C-prM gene junction sequences of the study strains with sequences collected from NCBI database representing diverse geographical regions of the world. Generalised time reversible model of nucleotide substitution with gamma distributed variations among sites were used. The robustness of tree was estimated using 1000 bootstrap replications under Nearest-Neighbor Interchange procedure with input distance determined by Maximum-Likelihood method [14].
Results
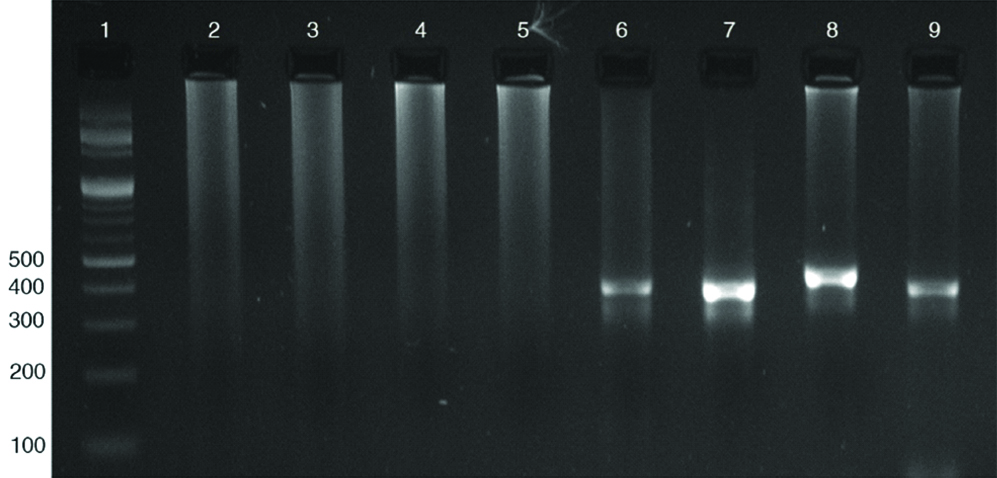
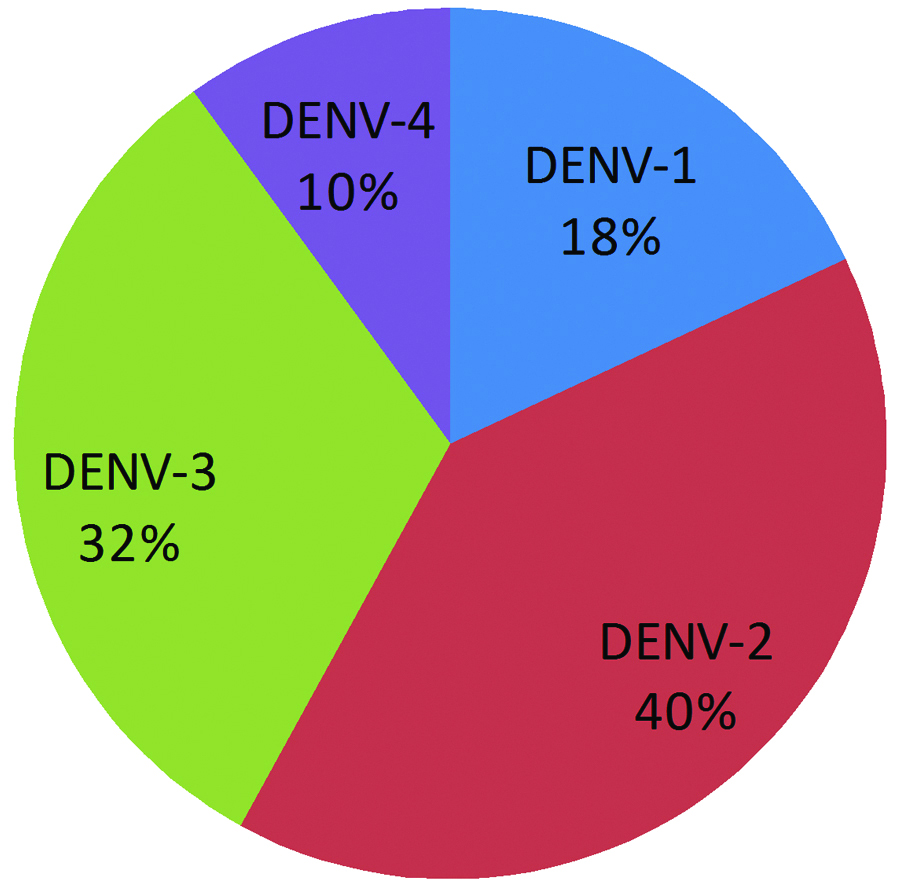
A total of 140 dengue positive samples were subjected to RT-PCR analysis to identify serotypes. Out of 140 samples, 45 samples were positive for different serotypes. Among the 45 PCR positive samples 12 (26.7%), and 5 (11.1%) were positive solely for Dengue virus-2 and 3 respectively while remaining 28 (62.2%) samples showed more than one serotype. The different combinations of serotypes observed in the present study are: one Dengue virus-1, 2 (2.2%), seven Dengue virus-2,3 (15.5%), 10 Dengue virus-1,2,3 (22.2%), three Dengue virus-2,3,4 (6.7%), and seven samples (15.6%) showed infection with all four Dengue virus serotypes. [Table/Fig-2] Representative gel picture showing the combined serotype infection with all four serotypes. [Table/Fig-3] depicting the serotype distribution among the PCR positive samples. The predominant serotypes were Dengue virus-2 and 3 when multiple serotypes were concerned [Table/Fig-4]. A single case of dengue virus-5 was reported in Malaysia during 2007 but the genome sequence was not yet published [2], hence the present study was limited to Dengue virus 1 to 4.
Representative gel picture showing the combined serotype infection with all four serotypes. Lane1-100 bp marker, lane 2-5 sample showing negative for all four serotypes, lane 6-9 sample showing positive for all four serotypes Dengue virus 1-411 bp, Dengue virus 2-403 bp, Dengue virus 3 453 bp and Dengue virus 4 401 bp.
Serotype distribution among the PCR positive samples (n=45).
Percentage individual serotype prevalence among PCR positive samples n=45 (multiple serotype infections were also included for percentage calculation).
Sequences obtained from the study were uploaded in the NCBI BLAST Database and sequence homology with different sequences was observed. Dengue virus-1 sequences showed 99% homology with sequences of Kerala, Sri Lanka, Thailand, Indonesia, Vietnam and Cambodian strains. Dengue virus -2 sequences showed 98% -100% homology with Kerala, Lucknow, Delhi, Odisha, Manipal, Arunachal Pradesh, Singapore and China strains. Dengue virus-3 sequences showed 99%-100% homology with Delhi, Orissa, Pune, Surat, Rajasthan, Kerala, China, Singapore and Pakistan strains.
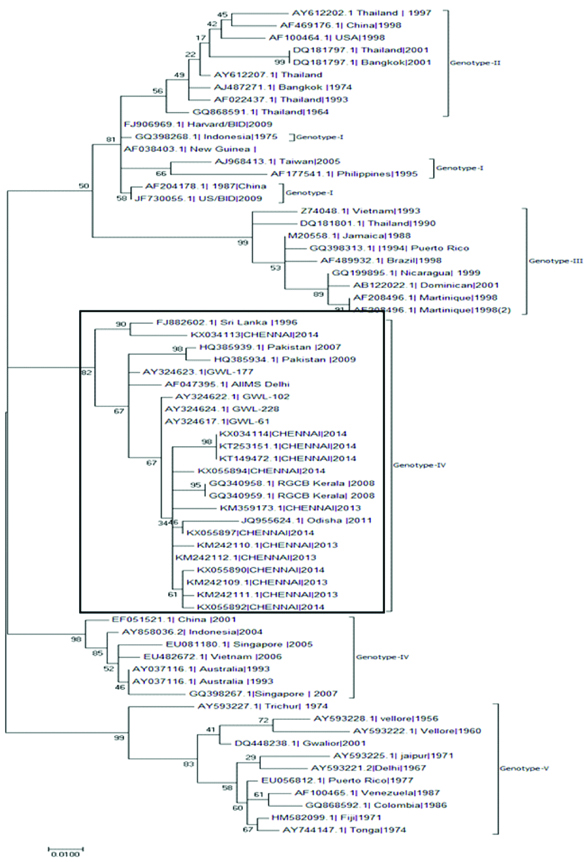
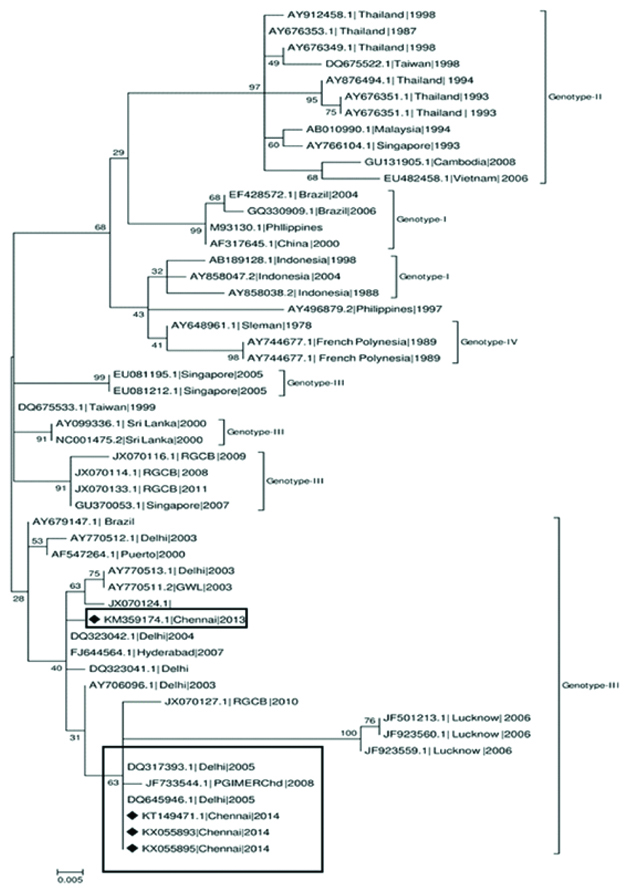
Phylogenetic analysis revealed that Dengue virus-1 sequences obtained from the present study belonged to genotype-I [Table/Fig-5], Dengue virus-2 sequences belonged to genotype-IV [Table/Fig-6], Dengue virus-3 belonged to genotype III and [Table/Fig-7]. The samples were contaminated during handling hence; authors were unable to perform the phylogenetic analysis for Dengue virus-4.
Dengue virus-1 Phylogenetic tree for current study sequences was constructed with sequences from diverse geographical regions. The tree was constructed using Maximum-Likelihood method by MEGA7 software.
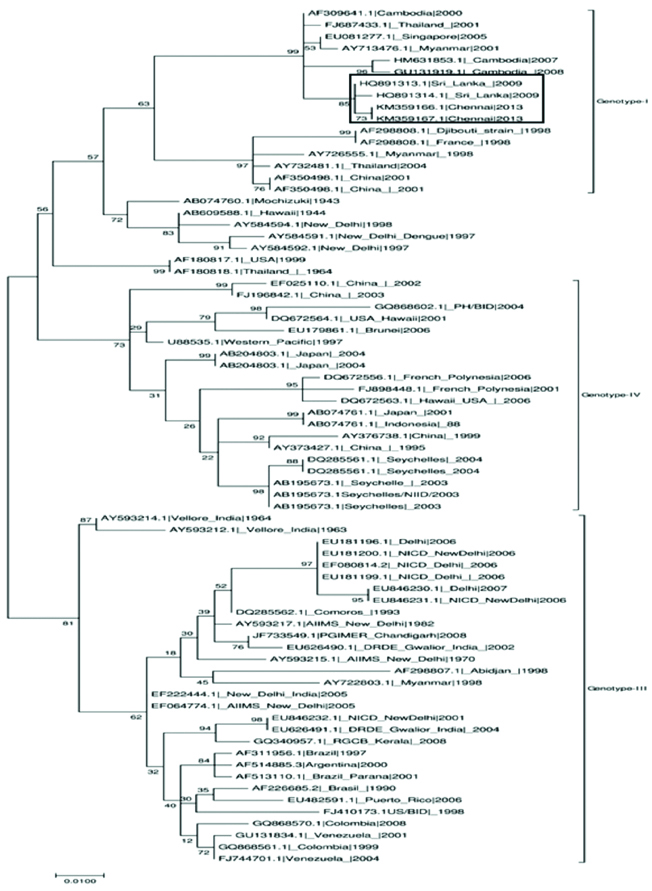
Dengue virus-2 Phylogenetic tree for current study sequences was constructed with sequences from diverse geographical regions. The tree was constructed using Maximum-Likelihood method by MEGA7 software.
Dengue virus-3 Phylogenetic tree for current study sequences was constructed with sequences from diverse geographical regions. The tree was constructed using Maximum-Likelihood method by MEGA7 software.
Discussion
Dengue fever is an important mosquito-borne arboviral infection all over the world [15]. Prevalence of dengue is predisposed by multiple factors such as the immune status of human hosts, age, virus traits, mosquito vector, environmental factors and human movement. Knowledge of different serotypes and genotypes in circulation is crucial for the development of an effective vaccine. In case of dengue virus, it is hindered by widespread circulation of four serotypes and different genotypes resulting in imperfect cross-protective immunisation [16].
Dengue virus serotypes and its genotype circulating in an area play a major role in the outcome of the disease and co-circulation of multiple serotypes along with their virulent genotypes is considered as the most important factor for severe fatal outbreaks in endemic regions [11]. Since the 1950’s, the co-circulation of all four-dengue virus serotypes is common in Southeast Asia and now it has expanded widely to tropics and subtropics of the world wherever the Aedes sp. mosquitoes are distributed [17].
Circulations of multiple serotypes with concurrent infections are being reported in most parts of the country and major outbreaks of DF and DHF are caused by Dengue virus-2 and 3 [18]. Multiple serotypes were reported in southern states of Kerala [19], Karnataka [20] and Telangana [21] nevertheless there was no study reporting the circulation of multiple serotype infections in Tamil Nadu. To the best of authors’ knowledge, this is the first study that observed the circulation of all four serotypes with different combinations of concurrent infections. Out of 45 PCR positive samples, 28 (62.2%) were shown five different serotype combinations of concurrent infection. The predominant serotypes detected in the present study are dengue virus 2 and 3 as recorded in Delhi [18,22-24], Kerala [25] and Karnataka [20].
The dengue virus 1 genotype-III is the predominant circulating genotype in India for the past six decades and the genotype-I circulated only during the year 1997-98 in Delhi which is the commonly circulating genotype in Southeast Asian countries [26]. In the present study, authors detected the circulation of genotype-I in Chennai. Shanthi G et al., reported the circulation of genotype-III [27]. Circulation of multiple genotypes and genotype replacement is a critical factor for the dengue management strategies due to the differences in the virulence between the genotypes [18]. Phylogenetic analysis of the sequences revealed clustering of the present study strains with Sri Lankan strains which were in circulation during the 2009 dengue epidemic [28].
Authors have detected the circulation dengue virus 2 genotype-IV in Chennai. Dengue virus-2 genotype IV is the predominant serotype in India [29,30] and was involved in major outbreaks that occurred in northern India. These serotypes were reported to be in continuous circulation with high capabilities to re-emerge and cause increased incidence of DHF [31]. The phylogenetic sequence analysis revealed clustering of the present study strains from Chennai with Delhi, Gwalior, Odisha, Kerala, Pakistan and Sri Lankan isolates.
Dengue virus-3 serotype detected in the present study belonged to genotype-III and clustered with Delhi and Gwalior sequences in the phylogenetic tree. This genotype of dengue virus- 3 is also circulated in most parts of India and caused major outbreaks during the past decade [32]. Circulation of dengue virus-3 genotype III has been reported in Madurai by Paramasivan R et al., 2010 [10].
Conclusion
Increased global travel contributes to the emergence, re-emergence, and circulation of multiple serotypes in a geographical region. Circulation of multiple serotypes and concurrent infection with heterologous serotype causes the severe form of dengue diseases such as dengue hemorrhagic fever and dengue shock syndrome. In the present study, authors have detected the circulation of multiple serotypes with a high number of concurrent infections. Re-emergence and continuous circulation of genotypes were also detected in the present study. The burden of dengue disease in India and Chennai are under-reported. Detailed continuous molecular surveillance studies and stringent strategies based on the outcome of such studies are urgently warranted to reduce dengue-related mortalities in any given region. The major limitation of the present study was the unavailability of clinical data to correlate with the study findings. The future direction of the study was planned to sequence the complete genome of all four serotypes to identify the novel significant mutations.
[1]. Klungthong C, Putnak R, Mammen MP, Li T, Zhang C, Molecular genotyping of dengue viruses by phylogenetic analysis of the sequences of individual genesJournal of Virological Methods 2008 154(1):175-81.10.1016/j.jviromet.2008.07.02118778736 [Google Scholar] [CrossRef] [PubMed]
[2]. Mustafa MS, Rasotgi V, Jain S, Gupta V, Discovery of fifth serotype of dengue virus (DENV-5): A new public health dilemma in dengue controlMedical Journal Armed Forces India 2015 71(1):67-70.10.1016/j.mjafi.2014.09.01125609867 [Google Scholar] [CrossRef] [PubMed]
[3]. Costa RL, Voloch CM, Schrago CG, Comparative evolutionary epidemiology of dengue virus serotypesInfection, Genetics and Evolution 2012 12(2):309-14.10.1016/j.meegid.2011.12.01122226705 [Google Scholar] [CrossRef] [PubMed]
[4]. Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Refining the global spatial limits of dengue virus transmission by evidence-based consensusPLoS Negl Trop Dis 2012 6(8):e176010.1371/journal.pntd.000176022880140 [Google Scholar] [CrossRef] [PubMed]
[5]. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, The global distribution and burden of dengueNature 2013 496(7446):504-07.10.1038/nature1206023563266 [Google Scholar] [CrossRef] [PubMed]
[6]. National Vector Borne Disease Control Programme (http://www.nvbdcp.gov.in/den-cd.html) [Google Scholar]
[7]. Thomas L, Verlaeten O, Cabié A, Kaidomar S, Moravie V, Martial J, Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 co-epidemicThe American Journal of Tropical Medicine and Hygiene 2008 78(6):990-98.10.4269/ajtmh.2008.78.99018541782 [Google Scholar] [CrossRef] [PubMed]
[8]. Chakravarti A, Arora R, Luxemburger C, Fifty years of dengue in IndiaTransactions of the Royal Society of Tropical Medicine and Hygiene 2012 106(5):273-82.10.1016/j.trstmh.2011.12.00722357401 [Google Scholar] [CrossRef] [PubMed]
[9]. Bhuvaneswari C, Raja R, Arunagiri K, Mohana S, Sathiyamurthy K, Krishnasamy K, Gunasekaran P, Dengue epidemiology in Thanjavur and Trichy district, Tamilnadu-Jan 2011-dec 2011Indian Journal of Medical Sciences 2011 65(6):26010.4103/0019-5359.10702823391835 [Google Scholar] [CrossRef] [PubMed]
[10]. Paramasivan R, Dhananjeyan KJ, Leo S, Muniaraj M, Thenmozhi V, Rajendran R, Dengue fever caused by dengue virus serotype-3 (subtype-III) in a rural area of Madurai district, Tamil NaduIndian J Med Res 2010 132:339-42. [Google Scholar]
[11]. Das B, Das M, Dwibedi B, Kar SK, Hazra RK, Molecular investigations of dengue virus during outbreaks in Orissa state, Eastern India from 2010 to 2011Infection, Genetics and Evolution 2013 16:401-10.10.1016/j.meegid.2013.03.01623523598 [Google Scholar] [CrossRef] [PubMed]
[12]. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV, Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reactionJournal of Clinical Microbiology 1992 30(3):545-51. [Google Scholar]
[13]. Fatima Z, Idrees M, Bajwa MA, Tahir Z, Ullah O, Zia MQ, Serotype and genotype analysis of dengue virus by sequencing followed by phylogenetic analysis using samples from three mini outbreaks-2007-2009 in PakistanBMC Microbiology 2011 11(1):20010.1186/1471-2180-11-20021906394 [Google Scholar] [CrossRef] [PubMed]
[14]. Kumar S, Stecher G, Tamura K, MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasetsMol Biol Evol 2016 33(7):1870-74.10.1093/molbev/msw05427004904 [Google Scholar] [CrossRef] [PubMed]
[15]. Weaver SC, Vasilakis N, Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral diseaseInfect Genet Evol 2009 9(4):523-40.10.1016/j.meegid.2009.02.00319460319 [Google Scholar] [CrossRef] [PubMed]
[16]. Raghwani J, Rambaut A, Holmes EC, Hang VT, Hien TT, Farrar J, Endemic dengue associated with the co-circulation of multiple viral lineages and localized density-dependent transmissionPLoS Pathog 2011 7(6):e100206410.1371/journal.ppat.100206421655108 [Google Scholar] [CrossRef] [PubMed]
[17]. Chinnawirotpisan P, Mammen MP, Nisalak A, Thaisomboonsuk B, Narupiti S, Thirawuth V, Detection of concurrent infection with multiple dengue virus serotypes in Thai children by ELISA and nested RT-PCR assayArchives of Virology 2008 153(12):222510.1007/s00705-008-0249-919011729 [Google Scholar] [CrossRef] [PubMed]
[18]. Chakravarti A, Chauhan MS, Kumar S, Ashraf A, Genotypic characterization of dengue virus strains circulating during 2007-2009 in New DelhiArchives of virology 2013 158(3):571-81.10.1007/s00705-012-1522-523129129 [Google Scholar] [CrossRef] [PubMed]
[19]. Reddy MN, Dungdung R, Valliyott L, Pilankatta R, Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013–2015 in northern Kerala, IndiaPeer J 2017 5:e297010.7717/peerj.297028316881 [Google Scholar] [CrossRef] [PubMed]
[20]. Vinodkumar CS, Kalapannavar NK, Basavarajappa KG, Sanjay D, Gowli C, Nadig NG, Episode of coexisting infections with multiple dengue virus serotypes in central Karnataka, IndiaJournal of Infection and Public Health 2013 6(4):302-06.10.1016/j.jiph.2013.01.00423806706 [Google Scholar] [CrossRef] [PubMed]
[21]. Vaddadi K, Gandikota C, Jain PK, Prasad VS, Venkataramana M, Co-circulation and co-infections of all dengue virus serotypes in Hyderabad, India 2014Epidemiology & Infection 2017 145(12):2563-74.10.1017/S095026881700147928726595 [Google Scholar] [CrossRef] [PubMed]
[22]. Gupta E, Dar L, Kapoor G, Broor S, The changing epidemiology of dengue in Delhi, IndiaVirology Journal 2006 3(1):9210.1186/1743-422X-3-9217083743 [Google Scholar] [CrossRef] [PubMed]
[23]. Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, IndiaVirology Journal 2008 5(1):110.1186/1743-422X-5-118182120 [Google Scholar] [CrossRef] [PubMed]
[24]. Islam A, Abdullah M, Tazeen A, Afreen N, Deeba F, HussainNaqvi I, Detection of all the four serotypes of dengue virus in New Delhi, India during post monsoon season of 2015Indian Journal of Health Sciences and Care 2016 3(1):24-29.10.5958/2394-2800.2016.00005.5 [Google Scholar] [CrossRef]
[25]. Anoop M, Issac A, Mathew T, Philip S, Kareem NA, Unnikrishnan R, Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South IndiaIndian J Exp Biol 2010 48(8):849-57. [Google Scholar]
[26]. Kukreti H, Dash PK, Parida M, Chaudhary A, Saxena P, Rautela RS, Phylogenetic studies reveal existence of multiple lineages of a single genotype of DENV-1 (genotype III) in India during 1956-2007Virology Journal 2009 6(1):110.1186/1743-422X-6-119126194 [Google Scholar] [CrossRef] [PubMed]
[27]. Shanthi G, Purushothaman I, Rajarajan S, Phylogenetic analysis of Dengue Virus Serotype 1 Isolated from Clinically Suspected Pediatric Patients in Chennai, TamilnaduInternational Journal of Advanced Biotechnology and Research 2016 7(1):106-11. [Google Scholar]
[28]. Tissera HA, Ooi EE, Gubler DJ, Tan Y, Logendra B, Wahala WM, New dengue virus type 1 genotype in Colombo, Sri LankaEmerging Infectious Diseases 2011 17(11):2053-55.10.3201/eid1711.10189322099096 [Google Scholar] [CrossRef] [PubMed]
[29]. Kumar NP, Jayakumar PR, George K, Kamaraj T, Krishnamoorthy K, Sabesan S, Genetic characterization of dengue viruses prevalent in Kerala State, IndiaJournal of Medical Microbiology 2013 62(4):545-52.10.1099/jmm.0.052696-023288429 [Google Scholar] [CrossRef] [PubMed]
[30]. Sharma P, Mittal V, Chhabra M, Singh P, Bhattacharya D, Venkatesh S, Continued circulation of DENV-2 (genotype IV) in Delhi, IndiaBritish Microbiology Research Journal 2015 11:1-8.10.9734/BMRJ/2016/21728 [Google Scholar] [CrossRef]
[31]. Dash PK, Parida MM, Saxena P, Kumar M, Rai A, Pasha ST, Emergence and continued circulation of dengue-2 (genotype IV) virus strains in northern IndiaJournal of Medical Virology 2004 74(2):314-22.10.1002/jmv.2016615332281 [Google Scholar] [CrossRef] [PubMed]
[32]. Sharma S, Dash PK, Agarwal S, Shukla J, Parida MM, Rao PV, Comparative complete genome analysis of dengue virus type 3 circulating in India between 2003 and 2008Journal of General Virology 2011 92(7):1595-600.10.1099/vir.0.030437-021411675 [Google Scholar] [CrossRef] [PubMed]