HCV replication has been detected at extra-hepatic sites like macrophages, B-cells, T-cells, bone marrow cells, dendritic cells and platelets in chronic HCV (CHC) infected patients [1]. It has been observed in recent studies that platelets serve as carrier of HCV [2,3]. Further, treatment of chronic HCV infection with Direct-Acting Antiviral (DAA) agents is associated with high rates of Sustained Virologic Response (SVR) >90%. Various reasons for treatment failure are liver fibrosis progression, previous antiviral therapy, viral factors such as baseline viral load, evidence of Resistance Associated Substitutions (RASs) and persistence of virus at extra-hepatic site [4]. Platelet-associated HCV particles have been shown to cause hindrance on the efficiency of antiviral therapy [4].
The quantitation of HCV-RNA in platelets or leukocyte components has generated contradictory results [5-7]. With this information in background, we decided to explore the impact of presence of virus in platelet on DAA treatment. The present study was done to detect and quantify HCV RNA from the platelets of infected patients and to compare it with parallel plasma levels.
Materials and Methods
Study Population and Sample Collection
This is a prospective, cross-sectional study involving consecutive patients with chronic HCV infection (n=62) who were seropositive for HCV and were attending Outpatient Department or were admitted in the inpatient ward from August 2016 to January 2017. Patients were excluded from the study if they had concurrent Hepatitis B virus or HIV infection. The antiviral treatment for HCV consisted of DAA ± Ribavirin as per the standard guidelines for 12 weeks [8]. Ribavirin was given if patient was found to be cirrhotic [9]. SVR was defined as undetectable serum HCV RNA 12 week post treatment [9]. The study was approved by the Institutional Ethics Committee (IEC) of the Institute, vide approval number IEC/2016/44/MA/14 in conformance to the ethical guidelines of the 1975 Declaration of Helsinki. Informed consent in writing was obtained from each patient prior to enrollment.
A total of 20 mL of whole blood in EDTA was collected at baseline for quantitation in plasma and platelet consequently at 4, 12, and 24 weeks post-treatment, viral load was monitored only in plasma to access treatment response.
Isolation of Platelets
Platelet Rich Plasma (PRP) was prepared by centrifuging whole blood sample at 300xg for 10 minutes. PRP was to obtain a platelet pellet. The pellet was washed seven times with Phosphate buffered saline. Leukocyte contamination was excluded by manual counting in Neubauer chamber. HCV RNA extraction was completed promptly using automated RNA extraction kit taking 0.8 mL of plasma and 50 μL of platelet pellets, the RNA is eluted in 200 μL Binding buffer and 50μL Proteinase K. Plasma was separated and stored at -80oC till further processing. Quantitation of HCV RNA was done by the Abbott Real Time HCV assay on the automated m2000sp/m2000rt platform with linear range of detection between log101.08 IU/mL to log108.00 IU/mL, according to manufacturer’s instructions [9,10].
HCV Genotyping
Samples with viral load >3 log10IU/mL were further processed for genotyping. As per the guidelines [10], genotype-specific treatment plan needs to be taken into consideration, as the antiviral activity of DAAs differs according to HCV genotype. 5’UTR region was amplified for genotyping.
In-house designed primer (forward primer 5’ATGGATCACTCCCCTGTGAGGAACT3’, reverse primer 5’GTCTACGAGACCTCCCGGGGCACT3’) was used. PCR was performed using Phusion high fidelity DNA polymerase (Thermoscientific Inc., Waltham, MA). Amplified products were purified by gel-excision using QIAquick gel extraction kit (QIAGEN, GmbH, Mannheim, Germany). Forward and reverse sequence reads were aligned and assembled using DNA Baser v3.5.1 (Heracle BioSoft SRL, Romania). Genotype assignment was done by comparing the obtained sequences with the reported sequences on the Basic Local Alignment Search Tool (BLAST) database of NCBI [11,12].
Statistical Analysis
Correlation between plasma and platelet HCV RNA load was calculated by Bland-Altman analysis. Intraclass correlation coefficient was calculated to find out the agreement between plasma and platelet HCV RNA load. Bivariate association and p-value was calculated by chi-square test. The p<0.05 was considered as statistically significant.
Results
The study included 62 CHC patients with antibody reactive to HCV. Most of the study participants were men (53.2%), with an age 45 (interquartile range, IQR: 22-85) years as shown in [Table/Fig-1]. Out of 62, 45 (72.5%) patients had detectable HCV RNA in plasma (viremic), while 17/62 (27.4%) were negative for HCV RNA (non-viremic) before treatment initiation as shown in [Table/Fig-2].
Baseline characteristics of the study population.
| Variable (n=62) | Number |
|---|
| Age (median, range) years | 45 (22-85) |
| Male: Female | 1.13:1 |
| Total bilirubin (median, range) mg*/dL | 1.05 (0.5-17.7) |
| Direct bilirubin (median, range) mg*/dL | 0.2 (0.1-10.3) |
| Aspartate aminotransferase (AST) (median, range) IU/mL† | 53.5 (22-1143) |
| Alanine aminotransferase (ALT) (median, range) IU/mL† | 55.5 (17-1611) |
| Cirrhotic, n (%) | 16 (25.8) |
| Median plasma RNA levels (log10 IU‡/mL) (range) | 5.7 (3.55-6.7) |
| Median platelet RNA levels (log10 IU‡/mL) (range) | 4.13 (0-5.5) |
*mg: Milligram; †ml: Milliliter; ‡IU: International Unit
Comparison of HCV RNA levels between plasma and platelet.
| | Plasma | Total |
|---|
| >12 IU*/mL† | Negative | |
|---|
| Platelet | >12 IU*/mL† | 43 | 9 | 52 |
| Negative | 2 | 8 | 10 |
| Total | | 45 | 17 | 62 |
*IU: international unit; †ml: Milliliter
In viremic, parallel platelet sample were positive in 43/45 (95.5%) patients. Overall, in the viremic group, median baseline plasma viral load was log105.7 (IQR: log103.55-6.7) IU/mL and median baseline platelet viral load was log104.13 (IQR: log100.0-5.55) IU/mL [Table/Fig-1].
In non-viremic, 9/17 (52.9%) patients had detectable HCV RNA in the parallel platelet sample [Table/Fig-2]. Seventeen out of 42 patients showed RVR while SVR after 24 weeks was observed in 39/42 patients. Three patients were non-responders as they demonstrated detectable viral load after 12 week post treatment [Table/Fig-3]. In 42 patients where viral load was >3 log10IU/mL, 28 (66.6%) patients were of gt 3, while 14 (33.3%) patients were of gt 1 [Table/Fig-4].
Plasma HCV RNA load at different treatment points.
| Patient group, n=42 | Baseline plasma viral load range, IU*/mL† | Baseline platelet viral load range, IU*/mL† | Plasma viral load at 4 week range, IU*/mL† | Plasma viral load at 12 week range, IU*/mL† | Plasma viral load at 24 week range, IU*/mL† | p-value |
|---|
| RVR (4 week)‡, 17 | 5.7×103-5×106 | 69-9.9×103 | 0-<12 | 0 | 0 | 0.425 |
| ETR (12 week)#, 21 | 3.5×103-4.5×106 | 0-3.6×105 | 27-5.16×104 | 0 | 0 | 0.453 |
| SVR (24 week)$, 42 | 3.5×103-5×106 | 0-3.6×105 | 0-5.16×104 | 0 | 0-6.1×104 | 0.456 |
*IU: international unit; †ml: Milliliter; ‡RVR: Rapid virological response; #ETR: End-of-treatment response; $SVR (12): Sustained virological response.
p-value was calculated by chi-square test. p<0.05 was considered as statistically significant.
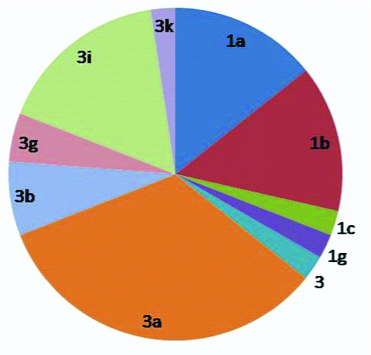
Genotype distribution of patients.
*1a(14.2%);1b(14.2%);1c(2.3%);1g(2.38%);3(2.38%);3a(33.3%);3b(7.14%);3g (4.76%); 3i (16.6%); 3k (2.38%); total genotype-1=28(66.6%); genotype-3=14(33.3%)
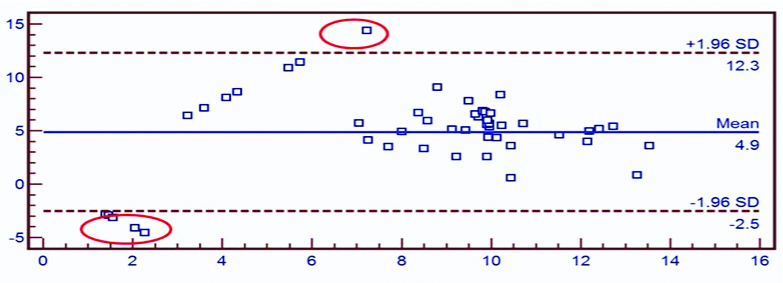
Overall the correlation between plasma and platelet viral load was 70.3%. Bland-Altman analysis showed 49/55(89%) agreement between plasma and platelet viral load, Intra-class Correlation Coefficient (ICC)=0.602, p-value <0.01, with concordance of 82.2%, sensitivity 95.5% and specificity of 47.05% [Table/Fig-5].
Level of agreement between plasma and platelet by Bland-Altman plot for all samples.
*X-axis: Mean viral load (log10 IU/mL); Y-axis: Difference between plasma and platelet viral load (log10 IU/mL)
Rapid Virological Response (RVR), Early Virological Response (EVR), End-Of-Treatment Response (ETR), and Sustained Virological Response (SVR) were observed in 38%, 30%, 35.7% and 92.8% respectively. A p-value was calculated by chi-square test between virological response and platelet viral load which was 0.425, 0.451, 0.453 and 0.456 for RVR, EVR, ETR and SVR respectively. Plasma and platelet viral load was found to be statistically insignificant (p=0.799).
In the bivariate unadjusted analysis, baseline platelet viral load did not influence treatment response. Non responders mean platelet viral load was log104.46 which was higher than responders (log103.27), though this association was statistically insignificant (p=0.456).
Discussion
Hepatocytes are the primary target cells of HCV; however HCV-RNA has been detected in other cells like monocytes, lymphocytes, dendritic cell and platelets [1]. Viruses like HIV, EBV, CMV and HCV etc., have been shown to directly interact with platelets and modulate their function [2] HCV RNA has been shown to remain stable and protected from degradation in platelets [3].
Similar to Santo M et al., Amer A et al., and Padovani JL et al., [5-7], all our patients harbor HCV-RNA in their platelets, independent of baseline viral loads. HCV-RNA levels were higher in plasma than in platelets, regardless of time of antiviral therapy, which was in agreement with previous studies. Given the importance of measuring viremia in the monitoring of HCV therapy, one concern is to select the best reservoir of virion. Differences in the sensitivity of viral load assays in different cell types, especially for low amounts of HCV RNA could affect the decisions regarding treatment and further patient management [5]. More reliability on monitoring the viral load in platelet pool of patients on Interferon based therapy has been observed [6]. This confirms that platelets may serve as reservoirs of HCV and helps in immune escape [7].
Limitation and Future Direction
This is the first study to study viral load in platelet pool for treatment monitoring in patients on DAA based therapy. However, small sample size of the study and lack of follow-up of patients in terms of achieving SVR was a major limitation. Larger studies need to be undertaken to confirm these observations and the usefulness of this reservoir for assessment of responses to treatment.
Further, cell-cell association studies are needed to study the mechanism of virion entry and replication within platelets.
Conclusion
HCV-RNA was efficiently detected in platelets. Our data reconfirms higher HCV-RNA levels in plasma compared to platelet, independent of time point of antiviral therapy. Viral load in platelet pool was higher in non-responders when compared to responders who achieved SVR.
Highlight
This study reiterates platelet as an extra hepatic reservoir of HCV, in addition to monocytes, lymphocytes and dendritic cell. The influence of HCV persistence in platelets among patients on DAAs and its effect on relapse of infection is also looked into in this study. Further, cell-cell association studies are needed to study the receptors involved in entry of HCV RNA in platelets. Whether compartmentalization of HCV in platelets is one of the factors responsible for persistence/resistance of virion to DAA therapy remains to be resolved.
*mg: Milligram; †ml: Milliliter; ‡IU: International Unit
*IU: international unit; †ml: Milliliter
*IU: international unit; †ml: Milliliter; ‡RVR: Rapid virological response; #ETR: End-of-treatment response; $SVR (12): Sustained virological response.
p-value was calculated by chi-square test. p<0.05 was considered as statistically significant.