The evolution and spread of antibiotic resistance is now recognised as a major health hazard and prudent use of antibiotics have been recommended by CDC and WHO. As such it remains important to continue surveillance of resistance patterns and mechanisms among human pathogens. Antibiotic resistance patterns and mechanisms in S. pneumoniae is being studied by many national and international groups [1-3]. However, the data regarding resistance patterns is highly variable and patterns change geographically. In India, a few studies have reported the antimicrobial resistance patterns [4-9], and in some regions, the data is more than two decades old. In our geographical region (Puducherry and its surroundings), studies have been conducted two decades ago [10]. The major determinants of macrolide resistance in pneumococcus were efflux–mediated (mefA/E) or target modification by enzymes (ermB). The relative prevalence of these genes varies among the regions and is an important factor in determining the levels of resistance to macrolides. The resistance to tetracycline is mediated solely be the tetM gene. The lack of data on genetic mechanisms of resistance in this region so-far needs to be addressed. Hence, it is necessary to determine the antibiotic resistance patterns and mechanisms in this region.
The pattern of serotype prevalence was found to be complex when compared across various regions in some multicentric studies [11,12]. The plan of introducing pneumococcal vaccination into universal immunisation programme in India was thus hindered by this epidemiological factor. Moreover, the colonisation rates, antimicrobial resistance/multidrug resistance and the treatment outcome varied among different serotypes [13]. Due to all these factors, studying pneumococcal serotype patterns continue to be a main area of study by many national and international groups.
The present study was hence, undertaken to study the antibiotic resistance patterns, mechanisms and its association with serotypes in our region.
Materials and Methods
Study description and ethics statement: This descriptive study was conducted from October 2014 to September 2017. The study was approved by institute human ethics committee (Approval No. JIP/IEC/2015/15/744). As the samples were sent for the routine diagnostic procedures, no separate consent was obtained from the participants. Demographic data was collected and recorded retrospectively, after the sample was positive for pneumococcus. The change in the trends of serotype distribution and antimicrobial resistance was determined based on a previously published study conducted at Jawaharlal Institute of Postgraduate Medical Education and Research from 1996 to 2000.
Bacterial isolates: Pneumococci isolated from various clinical specimens sent to the microbiology laboratory were identified by standard procedures like optochin susceptibility and bile solubility. For the confirmation of the isolates, polymerase chain reaction based on autolysin (lytA) gene was performed using published primers [14]. Isolates positive for lytA gene were further tested for the presence of cpsA gene using published primers and protocols [15]. Isolates which were positive for both the genes were tested for antimicrobial susceptibility profiles and capsular serotypes.
Antibiotic susceptibility testing and serotype deduction: All the isolates were tested for susceptibility to erythromycin (15μg), clindamycin (2μg), tetracycline (30 μg), levofloxacin (5μg) and trimethoprim-sulphamethoxazole (25μg) by Kirby-Bauer disc diffusion test (a suspension equivalent to 0.5 McFarland was inoculated onto Mueller-Hinton agar with 5% sheep blood and incubated in candle jar at 37°C for 18 hours) and the zone diameters of inhibition were interpreted based upon CLSI–2014 recommendations [16]. After testing for antimicrobial sensitivity, the isolates were stocked in STGG (Skimmed Milk-Trypticase Soy-Glucose-Glycerol) medium and preserved at -80°C. Minimum Inhibitory Concentration (MIC) for penicillin was determined by E-test using commercially available strips (Biomerieux, France). For all the antibiotic susceptibility tests, standard strain, Streptococcus pneumoniae ATCC 49619 was used as a control. Genetic mechanisms of macrolide and tetracycline resistance were detected by a multiplex PCR targeting ermB and mefA/E (for macrolide resistance) and (tetM, tetO, tetK and tetL) for tetracycline resistance separately using previously published primers [Table/Fig-1] [17]. Serotyping was performed by sequential multiplex PCR using primers and protocols mentioned in CDC laboratories (cpsA gene was used as an internal control and upto four primer pairs were included in each run) (supplementary materials S1 and S2) [18,19]. Initially, 25 isolates were serotyped at reference laboratories and these were subsequently used as controls for the multiplex PCR. The serotypes/groups were classified based upon the PCR-product sizes that are specific for the serotypes included in the run [Table/Fig-2]. All the PCR-products were run on 3% agarose gels stained with ethidium bromide and visualised under UV (GelDoc XR+, BioRad, USA).
Primers used for identification of antibiotic resistance genes.
| Resistance | Target | Forward primer* | Reverse primer* | Product size in bp |
|---|
| Macrolide resistance | erm(B) | TGGTATTCCAAATGCGTAATG | CTGTGGTATGGCGGGTAAGT | 745 |
| mef(A/E) | CAATATGGGCAGGGCAAG | AAGCTGTTCCAATGCTACGG | 317 |
| Tetracycline resistance | tet(M) | GTGGACAAAGGTACAACGAG | CGGTAAAGTTCGTCACACAC | 406 |
| tet(O) | AACTTAGGCATTCTGGCTCAC | TCCCACTGTTCCATATCGTCA | 515 |
| tet(K) | GATCAATTGTAGCTTTAGGTGAAGG | TTTTGTTGATTTACCAGGTACCATT | 155 |
| tet(L) | TGGTGGAATGATAGCCCATT | CAGGAATGACAGCACGCTAA | 229 |
* All the primer sequences are shown in 5’– 3’ direction
Serotype/groups included in each reaction of multiplex PCR.
| Reaction Number | Serotypes/groups included |
|---|
| 1 | 14, 1, 5, 4, 18A/B/C/F* |
| 2 | 6, 19F, 23F, 25F/25A/38*, 9V/9A* |
| 3 | 7B/7C/40*, 3, 15B/15C*, 7A/7F*, 17F |
| 4 | 8, 12F/12A/44/46*, 9N/9L*, 22F/22A*, 23A |
| 5 | 24F/24A/24B*, 2, 11A/11D*,19A, 16F |
| 6 | 21, 33A/33F/37*, 15A/15F*, 35F/47F*, 13 |
| 7 | 39, 23B, 35A/35C/42*, 20, 35B |
| 8 | 10F/10C/33C*, 34, 10A, 31 |
*The primers are common for the serotypes/groups mentioned
Statistical Analysis
The distribution of serotypes and non-susceptible isolates were expressed in percentages. Median MIC (MIC50) and MIC90 values and interquartile ranges were represented for penicillin. Chi-square test or Fischer-exact test were used as applicable to find any significant association between different variables and the significance of changes in MIC values were assessed using non-parametric Kruskal–Wallis test, with a two tailed p<0.05 considered as statistically significant. All the statistical analyses were done using IBM SPSS version 20.0 statistical software.
Results
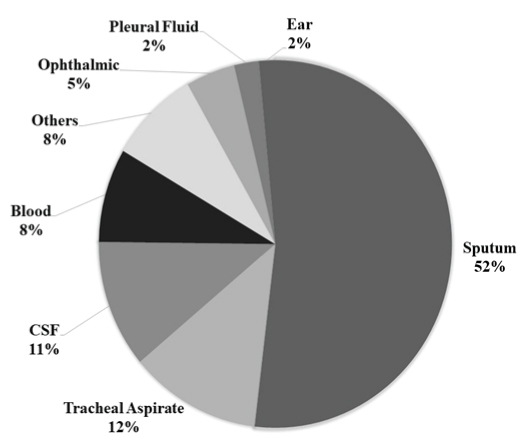
A total of 134 isolates collected over the study period were tested for their serotypes and antibiotic susceptibility profiles. The percentage of isolates from each type of sample is detailed in [Table/Fig-3].
Source wise distribution of pneumococcal isolates in the present study (n=134).
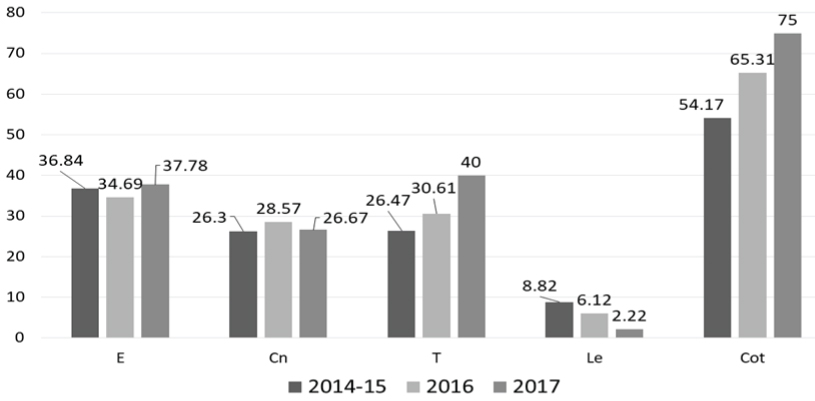
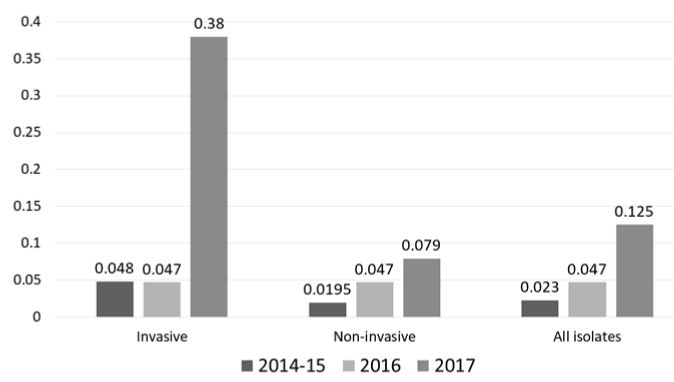
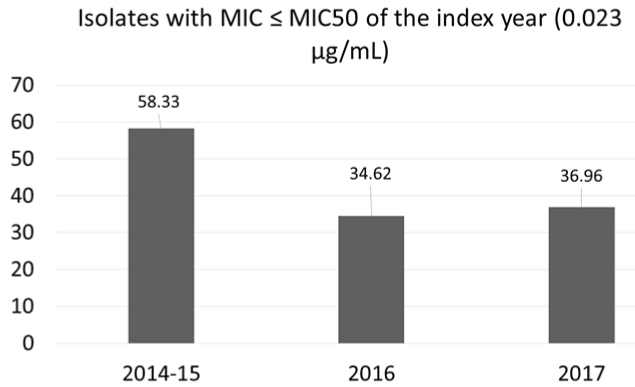
Antimicrobial Resistance Profile (AMR): Majority of the isolates were non-susceptible to trimethoprim-sulfamethoxazole (89/134; 66.4%), followed by erythromycin (47/134; 35.1%) and tetracycline (46/134; 34.3%) [Table/Fig-4]. Only two isolates were of the iMLSB phenotype. Resistance to fluoroquinolones (levofloxacin) was observed in only seven (5.2%) isolates. All the isolates were susceptible to vancomycin and linezolid. Resistance to tetracycline and trimethoprim-sulfamethoxazole increased from 26.5% and 54.2% in 2014 to 40% and 75% in 2017 respectively (p=0.757 and 0.218 respectively; Chi-square test), while resistance to levofloxacin decreased from 8.8% to 2.2% (p=0.678; Chi-square test). Of the 83 isolates tested for penicillin sensitivity, the MIC50 values for isolates from invasive and non-invasive sample types are shown in [Table/Fig-5]. MIC90 values for all the sample types in each year were 1.0, 0.5 and 1.5 μg/mL for the years 2014-15, 2016 and 2017 respectively. Only one isolate from sputum had MIC of 4 μg/mL, while eight isolates from CSF had MIC above 0.12 μg/mL (breakpoint for penicillin resistance for isolates causing meningitis) (MIC50=0.25 μg/mL, ranging 0.0598-0.473 μg/mL). There was an increase in MIC values across the three years [Table/Fig-5,6], suggesting a possible MIC creep. However, this shift in MIC50 was not significant (p=0.61; Kruskal–Wallis test). Around 14.9% (20/134) of the isolates were multidrug resistant, with macrolide-tetracycline-cotrimixazole being the most common profile.
Resistance to major antibiotics from 2014-15 to 2017.
All data shown in percentages. The total number of isolates tested is 134. E: Erythromycin; Cn: Clindamycin; T: Tetracycline; Le: Levofloxacin; Cot: Trimethoprim: sulphamethoxazole
Penicillin MIC50 trends across the three years of study.
MIC values were expressed in μg/mL
Percent isolates with MIC values less than MIC50 of the index year (0.023 μg/mL).
Relationship Between Serotype/Groups and AMR: Relationship between serotype/groups and AMR was statistically analysed only for the predominant serotype/groups and Non-Vaccine Types (NVT) (in total). Macrolide resistance was significantly associated with serotype/groups 6 (p=0.0005; Fischer-exact test) and 23 F (p=0.0003; Fischer-exact test), while resistance to trimethoprim-sulfamethoxazole was associated with serotypes 23F (p=0.04; Fischer-exact test) and 3 (p=0.009; Fischer-exact test). Resistance to penicillin, tetracycline, and levofloxacin were not significantly associated with any of the serotype/groups. All the NVT were susceptible to penicillin, the resistance rates to other antibiotics were low except for trimethoprim-sulfamethoxazole (61.9% were non-susceptible) and only one isolate was multidrug resistant among these isolates. Almost all of the MDR isolates were covered by PCV13 (80%) and PPV23 (90%).
Genetic mechanisms of resistance among serotypes: Among the 47 isolates resistant to erythromycin, 8(17%) had both ermB and mefA/E, while twenty two isolates (46.8%) had only ermB and sixteen isolates (34%) had only mefA/E as the genetic determinants of macrolide resistance. A significant association was found for the presence of ermB gene in serotype 23F (p=0.047; Fischer-exact test) and both ermB and mefA/E genes in serotype 19F (p=0.0014; Fischer-exact test). Of the seven isolates with both ermB and mefA/E, five isolates were of serotype 19F. Tetracycline resistance was mediated by tetM gene alone in all the tetracycline-resistant isolates.
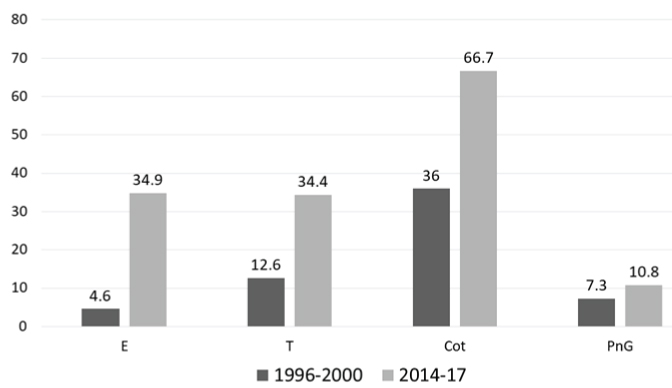
Trends in Serotype Distribution and Antimicrobial Resistance: Compared to 1996-2000, there was a significant increase in non-susceptibility to erythromycin (p<0.0001; Chi-square test), tetracycline (p=0.0015; Chi-square test) and trimethoprim-sulfamethoxazole (p<0.0001; Chi-square test), while penicillin resistance was not significantly different (p=0.46; Chi-square test) [Table/Fig-7]. When compared with the serotype pattern during the same period, serotypes/groups 1, 5, 7, 12 and have decreased, while 6, 14, 15, 19 and 23 have increased. There was a slight decrease in the vaccine coverage by both PCV13 and PPV23, with concomitant increase in the non-vaccine types.
Trends in antimicrobial resistance between 1996-2000 and 2014-17.
All data shown in percentages. The total number of isolates tested in 1996-2000 was 150 and in the present study were 134. E: Erythromycin; T: Tetracycline; Cot: Trimethoprim-sulphamethoxazole; PnG: Penicillin G
Discussion
Streptococcus pneumoniae with high rates of genetic recombination and natural transformation has impacted treatment options and vaccine programmes relevant to public health. Owing to the high plasticity of the genome, the phenotypic characters like antimicrobial resistance, serotype distribution and virulence factors are in a continuous shift, and this property has created difficulty in treating and controlling pneumococcal infections. It has been estimated that as much as 30% of sequence divergence can be found between any two isolates [20]. Presence of large genetic diversity compounds attempts to identify a suitable target for vaccine or antibiotics. Antimicrobial resistance genes are known to be contained in mobile genetic elements that can lead to horizontal genetic transfer among various micro-organisms. Widespread use of antibiotics like trimethoprim-sulfamethoxazole, penicillin, macrolides like azithromycin and erythromycin and tetracycline led to the emergence of antimicrobial resistance by various mechanisms like drug efflux (macrolides), target site modification (penicillin) or developing alternate metabolic pathways.
Although resistance to antimicrobials is a consistent feature, the relative prevalence of resistance varies significantly across various geographic regions [Table/Fig-8] [1,7,9-11,21-27].
Antimicrobial resistance profiles observed in studies from India [1,7,9-11,21-27].
| Study period | Location | Ery* | Tet† | Lev‡ | Cot§ | Pen|| | Reference |
|---|
| 1993-97 | Multicentric | 4.2 | - | - | 56.3 | 1.3 | [21] |
| 1996-2009 (in phases) | Multicentric | 1.3-17.4 | - | 0 | - | 3.8-30.4 | [1] |
| 2011-15 | Multicentric | 39 | - | - | 85 | 8 | [7] |
| 2012-15 | Multicentric | 40 | - | 0 | 100 | 20 | [22] |
| 2013-15 | Multicentric | 32 | 45 | 7 | 48 | 11.2 | [11] |
| 1996-97 | Vellore | 8 | 32 | - | 44 | 16 | [23] |
| 1996-2000 | Puducherry | 4.6 | 12.6 | - | 36 | 7.3 | [10] |
| 2007-10 | New Delhi | 20 | 34.9 | - | 83.3 | 5 | [24] |
| 2007-11 | Vellore | 13.6 | - | 1.2 | 87.3 | 4.5 | [9] |
| 2007-13 | Vellore | 30 | - | 0 | 96 | 5 | [25] |
| 2009-11 | Bangalore | 12.5 | - | 5 | 77.5 | 35 | [26] |
| 2009-11 | Bangalore | 28 | - | 4 | 44 | 3 | [27] |
| 2014-17 | Puducherry | 34.9 | 34.4 | 5.5 | 66.7 | 10.8 | This study |
The percentage of intermediate susceptible and resistant isolates shown. Table depicts duration of the studies and their corresponding locations.
*Erythromycin; †Tetracycline; ‡Levofloxacin; § Trimethoprim – sulphamethoxazole; || Penicillin
In all the studies, most of the isolates were resistant to cotrimoxazole. This has been attributed to previous widespread use of cotrimoxazole for treating acute respiratory infections. The same pattern was found in the present study, followed by resistance to macrolides and tetracycline. Macrolide resistance has been gradually increasing in India and globally, even reaching up-to 75% [28]. This may be due to increased use of macrolides like azithromycin to treat not only respiratory infections, but several other infections too. Over the counter sales in developing countries like India has led to indiscriminate use and increase in resistance. Penicillin resistance was initially low (1.3% in 1993-97) and it peaked in 2009-11 (35%). When compared to a previous study from the present hospital during 1996-2000, the resistance rates have significantly increased for all the antibiotics except penicillin. However, it should be noted that during 1996-2000 the breakpoints for penicillin resistance were different, and the isolates had MIC 0.1–1 μg/mL.
In the present study, a difference in the MIC values was noted among invasive and non-invasive isolates [Table/Fig-5] where the invasive isolates had a higher MIC50 values. Although it is generally believed that invasive isolates are more susceptible to antibiotics, the difference can be attributed to the frequency of invasive isolates in the present study (only 21% of the isolates were invasive). It should be noted that the MIC values were not more than 1μg/mL and hence, it was only low level resistance. The decrease in the number of isolates with MIC values less than or equal to MIC50 of 2014-15 (0.023 μg/mL) shows the gradual development of non-susceptibility to penicillin. This warrants the judicious use of beta lactams like amoxicillin to treat common infections like pharyngitis. Resistance to levofloxacin was not a common finding in India, and hence it appears that using levofloxacin or moxifloxacin could be a viable alternative to treat non-meningitis pneumococcal infections especially pneumonia and other invasive infections. This approach can further reduce the selective emergence of resistance to other antibiotics like macrolides and penicillin.
Macrolide resistance is known to be mediated mainly by two mechanisms-target site modification mediated by ermB gene, and macrolide efflux mediated by mefA/E. The relative prevalence of these two genes varied significantly across geographic regions [29,30]. A few isolates may possess both ermB and mefA/E, which significantly increases the MIC values. Some studies reported a positive association between serotypes and macrolide resistance. This is because a few non-vaccine serotypes like 19A (not covered in the initial PCV7) are known to harbor a large number of antimicrobial resistance genes, particularly ermB and mefA/E for macrolide resistance. After the use of PCV7 in countries like the USA and other parts of the world, there was a high incidence of pneumococcal infections caused by 19A clone ST320 [31]. This might have contributed to the increase in gene frequencies of ermB and mefA/E, which then spread horizontally across geographic regions. In the present study serotypes 23F and 19F were associated with the presence of ermB alone and ermB + mefA/E respectively. Overuse and abuse of macrolides for upper respiratory tract may have contributed to the increased resistance rates observed.
Various methods for serotyping pneumococci – Quellung reaction and latex agglutination tests using type-specific antibodies and molecular methods like multiplex PCR and real-time PCR targeting the serotype-specific genes were established [32]. Although antigen-based assays are more specific, they are costly and labour-intensive. Furthermore, only fresh cultures of bacteria can be tested by using these assays. These limitations sometimes favour use of molecular assays particularly when a fresh culture is unavailable. In the present study, serotyping of a few isolates was performed in reference laboratories in Vellore and Bangalore. The laboratories utilise a combination of these assays to deduce the serotypes. Upon receiving the results, the DNA from these isolates was used as controls in the subsequent serotype deduction by multiplex PCR. Although this approach is error-prone (inability to resolve a few serotypes), it remains a good choice for serotyping in resource-poor centres.
In the present study, serogroups 6 and 19 increased, while serotypes/groups 1, 5, 7 and 12 decreased or were absent when compared with 1996-2000 period. This shift in prevalence of pneumococcal serotypes over time has been studied in some populations as an effect of vaccination. However, in India pneumococcal vaccination is yet to be implemented nation-wide, and use of pneumococcal vaccines for high-risk groups is not documented. Hence although there is a great shift in pneumococcal serotypes, the same cannot be attributed to vaccine usage. The shift in serotype patterns may be due to other factors like difference in rate of colonisation/invasion, rates of antimicrobial/multidrug resistance, etc. The present study hence stresses the importance of studying the physiology of various pneumococcal serotypes and their invasive/colonisation potential. Use of genomics or transcriptomics can help in this regard.
Limitation
The present study performed only disc diffusion for testing routine antibiotics. Determining the MIC for these antibiotics along with that of penicillin may have helped in understanding the level of resistance/non-susceptibility to these drugs. The use of multiplex PCR for deducing serotypes is not very specific and at times it uses primers common for a few serotypes. The vaccine coverage estimated by this approach may be higher than using other more specific methods like latex agglutination or Quellung reaction for identification of various serotypes.
Conclusion
There has been a marked shift in antimicrobial resistance profiles over two decades (1996-2000 to 2014-17) in a single geographic area, demonstrating a dynamic process of acquiring drug resistance in pneumococcus. Continuous monitoring of antibiotic use and antibiotic susceptibility patterns is required for the successful treatment of pneumococcal infections. Use of more specific serotyping assay like latex agglutination test is recommended to better understand the serotype distribution. Genome-wide studies may help in understanding dynamics of serotype and antimicrobial resistance patterns in a better way.
* All the primer sequences are shown in 5’– 3’ direction
*The primers are common for the serotypes/groups mentioned
The percentage of intermediate susceptible and resistant isolates shown. Table depicts duration of the studies and their corresponding locations.
*Erythromycin; †Tetracycline; ‡Levofloxacin; § Trimethoprim – sulphamethoxazole; || Penicillin