Evaluation of the Epigenetic Biomarker Bone Morphogenic Protein 3 for Colorectal Cancer Diagnosis
Hasan Ashoori1, Mohammad Ebrahim Ghamarchehreh2, Mahmood Tavallaei3, Shahla Mohammad Ganji4, Mostafa Hosseini5, Mohammad Zolfaghari6, Zahra Ghamarchehreh7, Farnaz Vahidian8
1 Human Genetics Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran.
2 Baqiyatallah Research Center of Gastroenterology and Liver Disease, Baqiyatallah University of Medical Sciences, Tehran, Iran.
3 Human Genetics Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran.
4National Institute for Genetic Engineering and Biotechnology, Tehran, Iran.
5 Human Genetics Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran.
6 Student Research Committee, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
7 Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran.
8 Department of Biology, Science and Arts University, Yazd, Iran.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Mahmood Tavallaei, Human Genetics Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran.
E-mail: mhmzolfaghari1389@gmail.com
Introduction
Colorectal Cancer (CRC) is a common type of cancer with a rising prevalence worldwide. Morbidity and mortality of CRC can be reduced by screening programs and early diagnosis. Currently used screening tests include fecal-based methods are non-invasive and inexpensive, however the sensitivity and specificity of these tests are not high enough.
Aim
To evaluat hypermethylation of Bone Morphogenic Protein 3 (BMP3) as a biomarker in early diagnosis of CRC.
Materials and Methods
A total of 96 individuals were enrolled in the present case-control study (59 CRC patients versus 37 healthy controls) and Methylation-Specific Polymerase Chain Reaction (PCR) was used to evaluate methylation status of the BMP3 gene in plasma samples and colonoscopy tissue biopsies.
Results
In plasma samples, 75% of CRC patients showed hypermethylation in the BMP3 gene versus only 30% of the controls. In colonoscopy tissue biopsies, BMP3 showed hypermethylation in 81.6% of CRC patients. Specificity and sensitivity of this gene in CRC diagnosis were found to be 76% and 66% respectively.
Conclusion
The results of the present study showed that BMP3 in combination with other genes could be used as a non-invasive and promising biomarker in screening and diagnosis of CRC.
BMP3 protein, Colorectal neoplasms, Early diagnosis, Methylation
Introduction
Colorectal Cancer is a common type of cancer in several areas of the world [1,2]. The incidence of CRC is increasing in Asian countries like other countries worldwide [3]. CRC is the third most common cancer in Iran with an increasing prevalence in the last decade [4,5]. A recent research in Iran showed that CRC has an increasing rate in prevalence particularly in younger people because of changes in their lifestyles [6].
Delay in diagnosis is one of the most important problems in management and treatment of cancer, so a wide variety of studies have been focused on finding new strategies for early detection. There are increasing evidences showing that morbidity and mortality of CRC can be reduced by targeted screening programs and early diagnosis [7,8]. Designing an appropriate and successful screening program depends on discovering reliable markers and methods [9]. Similar to other malignancies, genetic and epigenetic changes play a key role in CRC development. Recently, several molecular genetics and epigenetic markers have been introduced for diagnosis, prognosis, and treatment of the CRC [10,11].
Methylation is the most common epigenetic change in cancer. DNA methylation of several genes in CRC has been reported [12]. The hypermethylation of gene promoters such as APC, MLH1 and Sept 9 is well known through numerous studies [13,14]. A currently-recognised molecular panel in diagnosis and screening of CRC includes BMP3 gene. Promoter hypermethylation of BMP3 has been reported in different cancers [15]. BMP3 is a member of TGF-beta superfamily with cell growth suppression role in different types of cancer [16]. In the present study, hyper methylation of the BMP3 gene was evaluated in plasma samples of CRC patients and healthy controls by Methylation Specific Polymerase Chain Reaction (PCR) in Iranian population.
Materials and Methods
In the present case control study, 37 healthy controls (with no evidence of disease) and 59 patients with CRC (verified by histopathological analysis) were enrolled. All patients provided written informed consent and the project was approved by the Local Committee on Health Research Ethics (Baqiyatallah university of medical sciences and health services). Five millilitres of peripheral blood sample was collected from patients using K3EDTA vacutainer tubes and tissue biopsies were provided from colonoscopy centre of Baqiyatallah hospital, Tehran, Iran from 2011 to 2014. Healthy controls selected from patients referred to colonoscopy centre with no evidence of CRC or other cancers.
DNA extraction: DNA was isolated from 500 μL plasma using the QIAamp DNA Blood Mini Kit (Qiagen-Germany) and total DNA from tissue biopsies was extracted using DNeasy Blood and Tissue Kit (Qiagen-Germany). The concentration of the isolated DNA was quantified using Nanodrop Spectrophotometer (Maestrogen-USA).
Bisulfite treatment: DNA was treated with bisulfite to convert unmethylated cytosines to uracils before MSP, as described previously [17].
MS-PCR: Methylation-Specific PCR (MSP) was performed to determine methylation status of the BMP3 gene. Primer sequences for the methylated and unmethylated templates are shown in [Table/Fig-1]. PCR product size of BMP3 was 103 bp. Each PCR reaction mix consisted of a total volume of 30 μL containing 15 μL hot start taq master mix, 1 μM concentration of each primer, 1 μL DMSO, 3 μL bisulfite-modified DNA and 9 μL RNase free water.
Primers designed for methylated and unmethylated BMP3 gene.
| BMP3 gene | Sequence (5’-3’) |
|---|
| M-Forward primer | TTTAGCGTTGGAGTGGAGACGGCGTTC |
| M-Reverse Primer | CGCGACCGAATACAACGAAATAACGA |
| U-Forward primer | TTTAGTGTTGGAGTGGAGATGGTGTTTG |
| U-Reverse primer | AAACACAACCAAATACAACAAAATAACAA |
M: Methylated; U: Unmethylated
Statistical Analysis
Statistical analysis was conducted using the SPSS 20.0 software (version 20.0, SPSS Inc, Chicago, USA). The Fisher’s-exact test and chi-square test were applied to study the statistical relationships between methylation status and pathological or demographical results as well as evaluation of hypermethylation in patients. A p-value less than 0.05 was considered significant.
Results
Demographic profiles of patients in case and control groups are shown in [Table/Fig-2]. There was no significant relationship between age, gender, Gastrointestinal Disease (GID) history and cancer history in study groups.
Demographic features of study groups.
| Characteristics | Control (n=37) | CRC (n=59) | p-value |
|---|
| Age (Mean, years) | 60.6 | 62.2 | 0.73 |
| Gender |
| Male (%) | 21 (57%) | 31 (53%) | 0.236 |
| Female (%) | 16 (43%) | 28 (47%) | |
| GID history |
| Positive (%) | 6 (16%) | 23 (39%) | 0.082 |
| Negative (%) | 31 (84%) | 36 (61%) | |
| GIC history in family | | | |
| Positive (%) | 7 (19%) | 18 (30%) | 0.475 |
| Negative (%) | 30 (81%) | 41 (70%) | |
CRC: Colorectal cancer; GID: Gastrointestinal disease; GIC: Gastrointestinal cancer
Methylation status in plasma samples: As shown in [Table/Fig-3], 44 of 59 (75%) CRC patients verses 11 of 37 (30%) healthy controls showed hypermethylation in the BMP3 gene [Table/Fig-4].
Methylation status of BMP3 in study population.
| Group | Control (n=37) | CRC (n=59) | p-value |
|---|
| Positive, M (%) | 11 (30%) | 44 (75%) | 0.026 |
| Negative, U (%) | 26 (70%) | 16 (25%) | 0.033 |
| Total (%) | 37 (100%) | 59 (100%) | |
M: Methylated; U: Unmethylated; CRC: Colorectal cancer
BMP3 methylation Status in affected CRC patients using methylated and unmethylated primers on 3% agarose gel. Size of PCR product and the ladder: 103 and 100 bp (respectively).
M: Methylated; U: Unmethylated; 1-4: Patients; P.C: Positive control; N.C: Negative control
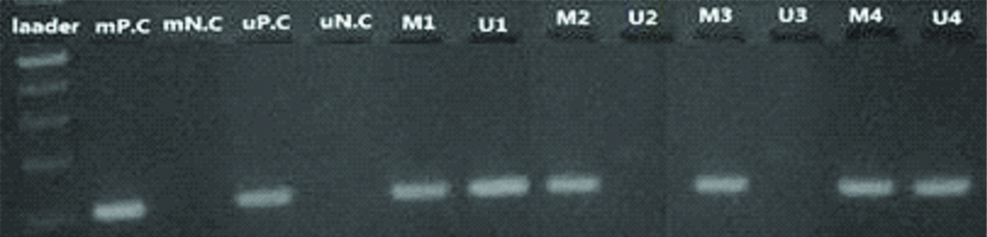
Specificity and sensitivity of BMP3: Evaluation of BMP3 methylation status in the study demonstrated that specificity and sensitivity of this gene for diagnosis of CRC was 76% and 66%, respectively.
Methylation status in colonoscopy tissue biopsies: Tissue biopsies were obtained from 30 CRC patients through the colonoscopy procedure. Methylation status of tissue biopsies was evaluated by MS-PCR. BMP3 was frequently hypermethylated in 81% of CRC patients.
Discussion
In the present study, BMP3 gene hypermethylation was evaluated in plasma and colonoscopy tissue biopsy samples of Iranian CRC patients versus healthy controls. All the patients had colorectal adenocarcinoma. In 74.5% of cases and 29.7% of controls, BMP3 showed hypermethylation. In a similar study, Houshmand M et al., evaluated BMP3 methylation status in CRC patients in Iran; their results showed that BMP3 has sensitivity and specificity about 57% and 93%, respectively [15]. In other countries, the methylation status of BMP3 was studied. Chen H et al., studied BMP3 methylation in tumour tissues of CRC patients and adenomas in China; results showed that BMP3 was hypermethylated in 66%, 74%, and 7% of CRC, adenomas and normal controls, respectively [18]. In another study carried out by Ashktorab H et al., methylation change was evaluated in seven CpG islands in the BMP3 gene promoter; results showed a significant difference in CRC as compared with the control group [19]. In Korea, Park SK et al., evaluated methylation of SFRP2, TFPI2, NDRG4, and BMP3 in stool DNA obtained from 111 patients. 40% of CRC samples and 33% of colorectal advanced adenoma samples showed hypermethylation in Korean population [20].
In this study, we used Methylation Specific PCR as a qualitative method for evaluating methylation status of the BMP3 gene. MSP is an inexpensive and easy method. In other studies different methods were used for assessment of hypermethylation such as methylation specific sequencing, High Resolution Melting (HRM) and array-based methods; accuracy and sensitivity of each method influence the results and validation of the results. Sensitivity and specificity of BMP3 in the detection of CRC were found to be 76% and 66%, respectively. It is suggested that BMP3 could be a valuable marker for CRC screening. However, it is recommended to use and compare this marker with other specific markers.
Limitation
One of the main limitations of the study was the sample size. The present study could be redesigned with a larger population of CRC patients to achieve more valid results.
Conclusion
Collectively, results of the present study indicated that BMP3 could be a promising biomarker for detection of colorectal adenocarcinoma and it seems to be a potential non-invasive marker.
M: Methylated; U: Unmethylated
CRC: Colorectal cancer; GID: Gastrointestinal disease; GIC: Gastrointestinal cancer
M: Methylated; U: Unmethylated; CRC: Colorectal cancer
[1]. Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasiaClin Gastroenterol Hepatol 2012 10(3):272-77.e1.10.1016/j.cgh.2011.10.00822019796 [Google Scholar] [CrossRef] [PubMed]
[2]. Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assayPLoS One 2008 3(11):e375910.1371/journal.pone.000375919018278 [Google Scholar] [CrossRef] [PubMed]
[3]. Salehi R, Atapour N, Vatandoust N, Farahani N, Ahangari F, Salehi AR, Methylation pattern of ALX4 genepromoter as a potential biomarker for blood-based early detection of colorectal cancerAdv Biomed Res 2015 4:25210.4103/2277-9175.17067726918234 [Google Scholar] [CrossRef] [PubMed]
[4]. Dolatkhah R, Somi MH, Bonyadi MJ, AsvadiKermani I, Farassati F, Dastgiri S, Colorectal cancer in Iran: molecular epidemiology and screening strategiesJ Cancer Epidemiol 2015 2015:64302010.1155/2015/64302025685149 [Google Scholar] [CrossRef] [PubMed]
[5]. Moamer S, Baghestani AR, Pourhoseingholi MA, Maboudi AA, Agha SH, Zali MR, Prognostic factors for survival in patients with colorectal cancer in iran between 2004-2015: competing risks regression analysis with generalized weibull modelBasic Clin Canc Res 2017 9(1):4-11. [Google Scholar]
[6]. Hessami Arani S, Kerachian MA, Rising rates of colorectal cancer among younger Iranians: is diet to blame?Curr Oncol 2017 24(2):e131-37.10.3747/co.24.3226 [Google Scholar] [CrossRef]
[7]. Barati Bagerabad M, Tavakolian S, Abbaszadegan MR, Kerachian MA, Promoter Hypermethylation of the Eyes Absent 4 Gene is a Tumour-Specific Epigenetic Biomarker in Iranian Colorectal Cancer PatientsActa Med Iran 2018 56(1):21-27. [Google Scholar]
[8]. Hu H, Chen X, Wang C, Jiang Y, Li J, Ying X, The role of TFPI2 hypermethylation in the detection of gastric and colorectal cancerOncotarget 2017 8(48):84054-65.10.18632/oncotarget.2109729137404 [Google Scholar] [CrossRef] [PubMed]
[9]. Garborg K, Colorectal cancer screeningSurg Clin North Am 2015 95(5):979-89.10.1016/j.suc.2015.05.00726315518 [Google Scholar] [CrossRef] [PubMed]
[10]. Singh MP, Rai S, Suyal S, Singh SK, Singh NK, Agarwal A, Genetic and epigenetic markers in colorectal cancer screening: recent advancesExpert Rev Mol Diagn 2017 17(7):665-85.10.1080/14737159.2017.133751128562109 [Google Scholar] [CrossRef] [PubMed]
[11]. Bedin C, Enzo MV, Del Bianco P, Pucciarelli S, Nitti D, Agostini M, Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patientsInt J Cancer 2017 140(8):1888-98.10.1002/ijc.3056527943272 [Google Scholar] [CrossRef] [PubMed]
[12]. Yi JM, Kim TO, Epigenetic alterations in inflammatory bowel disease and cancerIntest Res 2015 13(2):112-21.10.5217/ir.2015.13.2.11225931995 [Google Scholar] [CrossRef] [PubMed]
[13]. Matthaios D, Balgkouranidou I, Karayiannakis A, Bolanaki H, Xenidis N, Amarantidis K, Methylation status of the APC and RASSF1A promoter in cell-free circulating DNA and its prognostic role in patients with colorectal cancerOncol Lett 2016 12(1):748-56.10.3892/ol.2016.464927347211 [Google Scholar] [CrossRef] [PubMed]
[14]. Fu T, Liu Y, Li K, Wan W, Pappou EP, Iacobuzio-Donahue CA, Tumours with unmethylated MLH1 and the CpG island methylator phenotype are associated with a poor prognosis in stage II colorectal cancer patientsOncotarget 2016 7(52):86480-89.10.18632/oncotarget.1344127880934 [Google Scholar] [CrossRef] [PubMed]
[15]. Houshmand M, Abbaszadegan MR, Kerachian MA, Assessment of bone morphogenetic protein 3 methylation in Iranian patients with colorectal cancerMiddle East J Dig Dis 2017 9(3):158-63.10.15171/mejdd.2017.6728894518 [Google Scholar] [CrossRef] [PubMed]
[16]. Zhou X, Tao Y, Liang C, Zhang Y, Li H, Chen Q, BMP3 alone and together with TGF-β promote the differentiation of human mesenchymal stem cells into a nucleus pulposus-like phenotypeInt J Mol Sci 2015 16(9):20344-59.10.3390/ijms16092034426343641 [Google Scholar] [CrossRef] [PubMed]
[17]. Holmes EE, Jung M, Meller S, Leisse A, Sailer V, Zech J, Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urinePLoS One 2014 9(4):e9393310.1371/journal.pone.009393324699908 [Google Scholar] [CrossRef] [PubMed]
[18]. Chen W, Xiang J, Chen DF, Ni BB, Chen H, Fan XJ, Screening for differentially methylated genes among human colorectal cancer tissues and normal mucosa by microarray chipMol Biol Rep 2013 40(5):3457-64.10.1007/s11033-012-2338-923471507 [Google Scholar] [CrossRef] [PubMed]
[19]. Ashktorab H, Varma S, Brim H, Next-generation sequencing in African Americans with colorectal cancerProc Natl Acad Sci USA 2015 112(22):E285210.1073/pnas.150376011225941412 [Google Scholar] [CrossRef] [PubMed]
[20]. Park SK, Baek HL, Yu J, Kim JY, Yang HJ, Jung YS, Is methylation analysis of SFRP2, TFPI2, NDRG4, and BMP3 promoters suitable for colorectal cancer screening in the Korean population?Intest Res 2017 15(4):495-501.10.5217/ir.2017.15.4.49529142517 [Google Scholar] [CrossRef] [PubMed]