Introduction
Antimicrobial drug resistance is evolving as a serious threat to mankind due to indiscriminate use of antibiotics and lack of awareness about the mechanisms involved in drug resistance. Enterococcus faecalis, a common pathogen of the oral cavity has gained drug resistance over a period of years, making treatments refractory and ineffective.
Aim
To detect the antimicrobial resistance encoding genes of Enterococcus faecalis employing computational tools.
Materials and Methods:Antibiotic resistance genes were detected by retrieving sixteen genome sequences of Enterococcus faecalis from NCBI database which were further analysed using ResFinder. PlasmidFinder identified resistance encoding plasmids from recruited genomes under study. Protein sequences of most common phenotypes were subjected to protein BLAST and non-duplicate isolates showing 95-100% identity were selected for multiple sequence alignment using MEGA v7.0. Additionally, reconstruction of phylogenetic tree was performed to ascertain the diversity of these proteins among different genus and species.
Results
In silico analysis of genomes revealed that almost all the probed isolates exhibited resistance towards glycopeptides and macrolides. The genes lsa(A) were found in 100% of the isolates, followed by tet (M) and erm (B) exhibiting a frequency of 37.5% and 25% respectively. Apart from major antibiotics, the isolates also demonstrated resistance towards aminoglycoside, phenicol, tetracycline and trimethoprim class of antimicrobials.
Conclusion
The present investigation has emphasised the novelty in the application of in-silico tools in the understanding of the antibiotic resistance profile explored by the dreadful endodontic pathogen viz., E. faecalis. Further, this approach will aid in the implementation of improved treatment strategies and will facilitate to combat the dissemination of resistant gene cassettes to other oral pathogens or commensals residing in the vicinity.
Introduction
Enterococcus faecalis is a commensal and opportunistic pathogen commonly observed in oral diseases. It has been grouped as one among the 25 pathogens listed to be linked to persistent endodontic infections [1]. Interestingly, the frequency of oral carriage of E. faecalis remains inconsistent when considering different clinical conditions for example; individuals with gingivitis and periodontitis exhibit a prevalence of 3.7-35% [2], whereas it is 60% in diabetes patients and 6.6% in normal control group [3]. One of the studies on age and gender based carriage of E. faecalis showed 94.1% prevalence in children, 89.5% in adults and 81.6% in elderly Brazilian population. The study also reported that the predominant enterococcal species isolated from oral rinse was E. faecalis (88.7%) [4].
The pathogen is considered to be a microbial menace due to its frequent association with failure of treatments precipitated by resistance to antimicrobials [5]. They possess intrinsic resistant mechanisms and also have the ability to acquire gene cassettes coding for drug resistance through horizontal gene transfer. E. faecalis has been found to be associated with carious lesions, chronic periodontitis [6], persistent [7] and primary endodontic infections [8] along with a polymicrobial cosmos. Recalcitrant biofilm formation is often observed in both treated and untreated root canals [9]. Deep seated infections involve a polymicrobial community and anaerobic environment. Hence, drugs targeted against these pathogens may also pose a selective pressure on other organisms at the vicinity including E. faecalis which compels them to resist drugs and propagate in this stringent environment by modifying their genetic make-up or acquisition of drug resistant genes [10]. Several drug resistant isolates have been recovered from root canal infections [11,12].
The presence of such antibiotic resistant bacterial population does not only hamper the treatment process, but also acts as a reservoir for drug resistant genes which can be acquired by other organisms or even commensals residing in the vicinity of these pathogens. The transfer of such genes is enabled by mobile elements viz., plasmids and transposons [13].
The rationale behind this study is to project those interesting and rare drug resistant phenotypes exhibited by the pathogen. Hence, the basic drug resistant profile of E. faecalis needs to be validated to arrive at a conclusion about the novel mechanisms underlying resistance phenotype exhibited by this organism. The present study was aimed to probe into their genome for novel drug resistant genes and diversity of protein encoded by the predominant genes. To the best of our knowledge, this is the first study which reports acquired drug resistant genes in E. faecalis by in silico analysis.
Materials and Methods
Strain: A cross sectional study was designed to analyse sixteen whole genome sequences of Enterococcus faecalis retrieved from the NCBI database [14] as of Feb’ 2018, scaffolds and contigs were excluded. Also, the size of the genome, replicons, GC percentage, number of genes and proteins encoded were derived from the same database [14] [Table/Fig-1]. Assembled sequences were submitted to MLST (Multilocus sequence typing) 1.8 algorithm from Center for Genomic Epidemiology (CGE) [15]. MLST was performed including seven housekeeping genes (aroE, gdh, gki, gyd, pstS, xpt, yqiL) of E. faecalis to classify the strains based on the sequence types.
Strains of Enterococcus faecalis genome selected for the present study.
| Strain name/number | Sequence type | Size(Mb) | GC % | Replicons | Genes | Proteins |
|---|
| E.faecalis V583 | ST-6 | 3.36 | 37.3 | ChromosomePlasmids pTEF1, 2 | 3412 | 3264 |
| E.faecalis OG1RF | ST-1 | 2.74 | 37.8 | Chromosome | 2710 | 2602 |
| E.faecalis 62 | ST-66 | 3.13 | 37.4 | ChromosomePlasmid EP62pA | 3157 | 3075 |
| E.faecalis D32 | ST-40 | 3.06 | 37.4 | ChromosomePlasmids EFD32pA, B | 3174 | 2973 |
| E.faecalis-symbioflor 1 | ST-248 | 2.81 | 37.7 | Chromosome | 2885 | 2733 |
| E.faecalis DENG1 | ST-191 | 2.96 | 37.5 | Chromosome | 3050 | 2881 |
| E.faecalis ATCC 2912 | ST-30 | 3.04 | 37.3 | ChromosomePlasmids p1, p2 | 3128 | 2922 |
| E.faecalis LD33 | ST-25 | 2.80 | 37.6 | Chromosome | 2867 | 2695 |
| E.faecalis KB1 | ST-9 | 3.03 | 37.2 | Chromosome | 3014 | 2815 |
| E.faecalis L9 | ST-29 | 2.69 | 37.7 | Chromosome | 2706 | 2578 |
| E.faecalis L12 | ST-711 | 2.67 | 37.8 | Chromosome | 2660 | 2543 |
| E.faecalis CLB21560 | ST-28 | 3.24 | 37.8 | ChromosomePlasmids pA, pB | 3404 | 3211 |
| E.faecalissoralis | ST-65 | 3.05 | 37.2 | ChromosomePlasmids p1, p2 | 3059 | 2886 |
| E.faecalis W11 | Unknown | 2.70 | 37.7 | Chromosome | 2699 | 2577 |
| E.faecalis AR01/DG | ST-108 | 2.88 | 37.5 | ChromosomePlasmids ARO1.1, ARO1.2 | 2929 | 2788 |
| E.faecalis FDA ARGOS_338 | ST-19 | 2.86 | 37.6 | Chromosome | 2939 | 2572 |
Resistant Gene Profiling:Resfinder 3.0 is a user-friendly, computational tool [16] from the CGE. The whole genome sequence in the FASTA format was used as an input file to obtain facts about drug resistant genes present in the pathogen. The test was conducted for acquired antimicrobial resistance including all available antimicrobial drugs of six major classes such as aminoglycoside, beta-lactam, colistin, fluoroquinolone, fosfomycin, fusidic acid, glycopeptide, macrolide, lincosamide, streptogramin, nitroimidazole, oxazolidinone, phenicol, rifampicin, sulphonamide, rifampicin, trimethoprim and run using default parameters [Table/Fig-2].
Acquired antimicrobial drug resistant genes present in E.faecalis strains as detected by ResFinder.
| Strain name/number | Aminoglycoside | Glycopeptide | Macrolide, Lincosamide, Streptogramin B | Tetracycline | Phenicol | Trimethoprim |
|---|
| E.faecalis V583 | aac(6’) – aph (2”) | van X-B, van B, van H-B, van W-B,van Y-B, van S-B,van R-B | lsa(A)erm(B) | - | - | - |
| E.faecalis OG1RF | - | - | lsa(A) | - | - | - |
| E.faecalis 62 | - | - | lsa(A) | tet(M) | - | - |
| E.faecalis D32 | ant(6)Ia | - | lsa(A)erm(B) | | - | - |
| E.faecalis-symbioflor 1 | - | - | lsa(A) | | - | - |
| E.faecalis DENG1 | - | - | lsa(A) | tet(M) | - | - |
| E.faecalis ATCC 29212 | - | - | lsa(A) | tet(M) | - | - |
| E.faecalis LD33 | - | - | lsa(A) | | - | - |
| E.faecalis KB1 | - | - | lsa(A) | | - | - |
| E.faecalis L9 | - | - | lsa(A) | | - | - |
| E.faecalis L12 | - | - | lsa(A) | | fex(A) | |
| E.faecalis CLB21560 | aac(6’) – aph (2”)ant(6)Ia, aph(3’)-III | - | lsa(A)erm(B) | tet(M) | - | dfr(G) |
| E.faecalissoralis | - | - | lsa(A) | | - | - |
| E.faecalis W11 | - | - | lsa(A) | | - | - |
| E.faecalis AR01/DG | - | van Z-A, van Y-A,van X-A, van A,vanH-A, vans A, vanR-A | lsa(A)erm(B) | tet(L)tet(M) | - | - |
| E.faecalis FDA ARGOS_338 | - | - | lsa(A) | tet(M) | - | - |
E.faecalis FDA ARGOS_338
Plasmid Profiling: PlasmidFinder 1.3 is yet another effective tool from CGE to probe the presence of plasmids in the genomes selected [17]. The algorithm ran using default parameters and the reference database selected was that of Enterococcus, Streptococcus and Staphylococcus. The data obtained from the study was used to correlate between the presence of plasmids and the drug resistant phenotypes recorded earlier [Table/Fig-3].
Association between plasmids and resistant phenotypes.
| Strain name/number | Plasmids | % Identity | Resistant Phenotypes |
|---|
| E.faecalis V583 | pTEF1, 2, 3, pAD1 | 100 | Aminoglycoside, Glycopeptide, MLS |
| E.faecalis OG1RF | - | - | MLS |
| E.faecalis 62 | p703/5, pTEF2, pCF10 | 10099.596.4 | TetracyclineMLS |
| E.faecalis D32 | pGB354 | 96.6 | AminoglycosideMLS |
| E.faecalissymbioflor 1 | - | - | MLS |
| E.faecalis DENG1 | - | - | TetracyclineMLS |
| E.faecalis ATCC 29212 | pTEF3, pAD1 | 95.595.9 | TetracyclineMLS |
| E.faecalis LD33 | - | - | MLS |
| E.faecalis KB1 | - | - | MLS |
| E.faecalis L9 | - | - | MLS |
| E.faecalis L12 | - | - | Phenicol, MLS |
| E.faecalis CLB21560 | pAD1, pTEF2 | 10099.5 | Aminoglycoside, Tetracycline, Trimethoprim,MLS |
| E.faecalissoralis | pTEF2 | 96.7 | MLS |
| E.faecalis W11 | - | - | MLS |
| E.faecalis AR01/DG | p703/5,pAD1,pTEF3 | 10097.095.6 | Glycopeptide, Tetracycline,MLS |
| E.faecalis FDA ARGOS_338 | - | - | Tetracycline, MLS |
MLS: Macrolide; Lincosamide; Streptogramin B
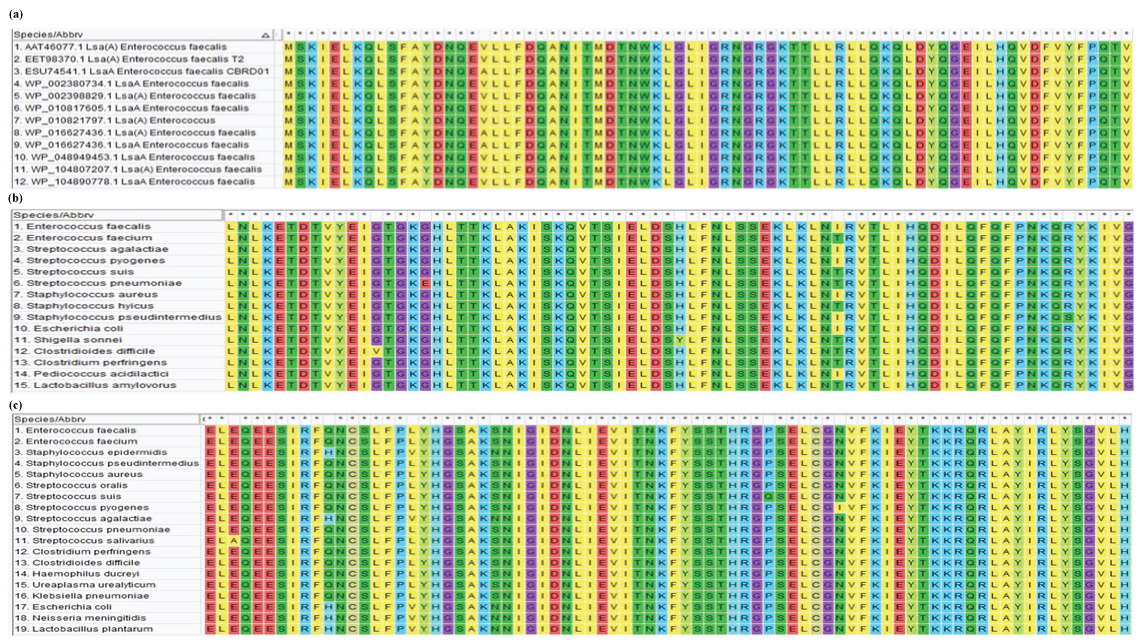
Multiple Sequence Alignment (MSA): The protein sequences of predominant drug resistant phenotypes coding for lsa(A), tet(M) and erm(B) were further analysed across several genus and species. Since lsa(A) was mostly present in Enterococcus spp. alone MSA was performed using FASTA sequence of different strains of E. faecalis. However, tet(M) and erm(B) were found to occur in diverse microbial population and hence, was subjected to MSA using MUSCLE programme of MEGA v.7.0 to ascertain the diversity of these proteins [18]. UPGMB was the clustering method applied with default parameters. Non-duplicate isolates exhibiting 100% query coverage with 95-100% identity were selected for the analysis [Table/Fig-4]. The subcellular localisation of the protein was deduced by PSORTb v3.0 [19].
Multiple sequence alignment demonstrating diversity of (a) Lsa(A), (b) Tet (M) protein and (c) Erm (B) protein among different bacterial genus and species.
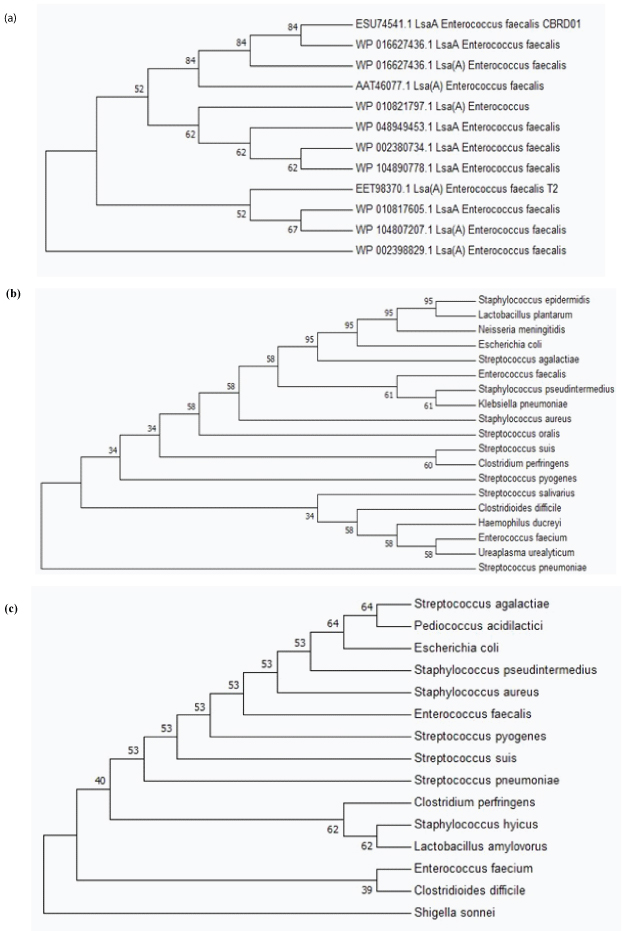
Phylogenetic Analysis of Erm(B) AND Tet(M) Protein: The diversity in course of evolution of LsaA, Erm(B) and Tet(M) proteins [Table/Fig-5] were inferred by using the Maximum Likelihood method based on the Poisson correction model [20]. Evolutionary analyses were conducted in MEGA v7.0 [21]. The bootstrap consensus tree inferred from 500 replicates was taken to represent the evolutionary history of the protein analysed [22].
Phylogenetic tree demonstrating the diversity of (a) Lsa (A), (b) Tet (M) and (c) Erm (B) protein among different bacterial genus and species.
Results
The principal objective of the present study was to acquire a basic understanding on the drug resistant profiles exhibited by E. faecalis strains submitted in public domain from across the globe. Sixteen whole genome sequences of E. faecalis returned resistance profiles as detected by ResFinder 3.0 corresponding to: 1) aminoglycoside; 2) glycopeptides; 3) macrolide, lincosamide streptogramin B (MLS); 4) tetracycline; 5) phenicol; and (6) trimethoprim. Among the six antimicrobial drug groups gene lsa(A) encoding MLS resistance was found to be the most predominant (n=16; 100%), followed by tet(M) (n=6; 37.5%) and erm(B) (n=4; 25%). Three strains out of sixteen harboured more than four resistance encoding genes (25%) [Table/Fig-6]. The multi-locus sequence typing showed that each of the strains belongs to a specific sequence type and they may not be clustered into the same sequence type.
Frequency of occurrence of antibiotic resistant genes in the strains of E.faecalis.
| Gene | Frequency (N=16) | Percentage (%) |
|---|
| lsa (A) | 16 | 100 |
| tet (M) | 6 | 37.5 |
| erm (B) | 4 | 25 |
| van group of genes | 2 | 12.5 |
| aac(6’)-aph (2”) | 2 | 12.5 |
| ant(6)Ia | 2 | 12.5 |
| aph(3’)-III | 1 | 6.25 |
| fex (A) | 1 | 6.25 |
| dfr (G) | 1 | 6.25 |
| tet (L) | 1 | 6.25 |
Plasmid profiling of the sequences retrieved were consistent with the data acquired from NCBI database. The plasmid type and % identity to the known plasmid sequences are provided in [Table/Fig-3]. Interestingly, aminoglycoside, glycopeptide and trimethoprim resistant phenotypes were found to be occurring only in strains harbouring plasmids, whilst the other phenotypes like tetracycline, phenicol and MLS were found in both plasmid bearing and non-plasmid containing strains [Table/Fig-3].
Multiple sequence alignment of Tet(M) and Erm(B) proteins showed conserved sequences among a diverse group of organisms tested [Table/Fig-4]. Single amino acid variations were dispersed along the protein sequences which indicates the process of evolution of genes encoding these proteins. The MSA for Lsa(A) protein which was species specific and conserved among Enterococci with variations dispersed throughout the protein sequence [Table/Fig-4]. The subcellular localisation of proteins as deduced by PSORTb v3.0 showed Lsa(A) to be in the transmembrane region and Tet(M) and Erm(B) proteins in the cytoplasm. Phylogenetic analysis performed using the protein sequences of Tet(M) and Erm(B) showed the evolution and diversification of protein among several genus and species analysed. Phylogenetic analysis of Lsa(A) protein showed diversity among the E.faecalis species studied, demonstrating rapid evolution of this protein at the species level [Table/Fig-5]. Thus, the present study provides an insight about the genetic basis of drug resistance exhibited by the strains of E. faecalis investigated.
Discussion
Antimicrobials are an integral part of the health care system and find its use in various fields of medicine including dentistry. The emergence of drug resistant pathogens has turned the treatment options towards the dark side. The World Health Organization (WHO) has reiterated the fact that treatment failure due to resurgence of drug resistant organisms will be the challenge faced in this millennium [23]. The commonly used drugs in dentistry are erythromycin, vancomycin, tetracycline, doxycycline, metronidazole etc., Although, local and systemic modes of delivery are used, local administration of antibiotics is preferred since the target tissue can be reached easily. This method also creates a selective pressure on the organisms infecting tissues, thus paving way for acquisition of drug resistant genes from similar pathogens. Since most of the drug resistant genes are carried on mobile elements like plasmids, transposons and integrons, the horizontal dissemination of drug resistant genes becomes undemanding.
The present study unravels the antibiotic resistant profiles of E. faecalis which is considered to be one of the vital pathogens related to endodontic and periodontal infections. Resistance towards vancomycin and tetracycline have been extensively studied in this pathogen. Interestingly, we could observe a few rare genes such as fex(A) and dfr(G) in two of the strains of E. faecalis (E.faecalis L12, E.faecalis CLB21560). Most of the Gram positive organisms exhibit resistance towards MLS class of drugs via two major mechanisms viz., drug efflux and demethylation of 23S rRNA. The gene lsa(A) is responsible for intrinsic resistance to lincosamides and streptogramins A in E. faecalis [24]. BLAST analysis and several other reports confirmed the species specificity of this protein. Although the role of Lsa remains ill-defined it may be involved in drug efflux pathway, as the protein resembles ATP-Binding Cassette (ABC)-efflux pumps. Clindamycin and quinupristin-dalfopristin resistance was reported in 100% of the strains exhibiting this phenotype. Targeted inactivation of lsa(A) gene produced susceptible phenotypes, while recombinant plasmid carrying lsa gene mediated complementation restored the resistant phenotype [25].
Another best understood mechanism of streptogramin resistance is through the demethylation of 23S rRNA [26]. The Erm proteins disrupt the binding between macrolides and rRNA by the transfer of methyl group at A2058 of 23S rRNA rendering the strains resistant to MLS. erm(A) and erm(B) which confers MLS(A) and MLS(B) phenotypes are carried on broad host range plasmid such as pAMβ1 [27]. This report is in agreement with the present study where we could observe erm (B) gene in plasmid bearing strains of E. faecalis. Hospital strains of E. faecalis have recorded highest prevalence of resistance to erythromycin (34.1%) which is mostly encoded by erm(B) gene (70.9%) [28]. Erm type of protein shows diversity across different genus and species, thus elucidating an underlying genetic evolution of these species. Since erythromycin is routine drug of choice in dental clinics, periodical monitoring and investigations into resistance genes encoding Erm protein should be performed in E. faecalis isolated from tissue or oral site which is non-compliant with treatment. This may reduce further complications by resistant species and also prevent horizontal gene transfer among other organisms in oral cavity.
Tetracycline resistance in Enterococci dates back to early 1960, soon after its introduction in clinical settings. The most prevalent resistant genes isolated from asymptomatic apical periodontitis were reported to be tet(M) (42%), tet(W) (29%) and erm(C) (24%) [29]. In vitro resistance studies on plaque samples demonstrated multidrug resistance in E. faecalis strains. The resistance profile in decreasing order was clindamycin and metronidazole (100%), erythromycin (80.8%) and tetracycline (53.2%) [30]. The Tet(M) protein mediates resistance via ribosomal protection while, Tet(L) confers resistance through drug efflux mechanisms which are energy dependent [31]. The present study also reports an overall prevalence of 43.8% of tetracycline encoded genes which is in close association with other in vitro studies.
Gentamycin resistance is generally mediated by Aminoglycoside Modifying Enzymes (AME).
The aac(6’) – aph (2”) possess both adenyltransferase and phosphotransferase activities, which confers resistance to gentamycin, amikacin, tobramycin and kanamycin. The encoded enzyme is carried on transposons accommodated within the plasmid or the chromosomal DNA. Interestingly, aac(6’)-aph (2”) was also observed in strains harbouring plasmids in the present study, which validates the location of this gene in the pathogen. These mobile elements also facilitate transfer of gene clusters to other anaerobic pathogens found in deep seated wounds. An investigation on the distribution of AME phenotype conducted in Japan detected aac(6’)-aph (2”) in about 42.5% of E. faecalis strains [32]. Strains with aac(6’)-aph (2”) +ant(6)-Ia + aph(3’)-IIIa phenotype showed high level resistance to gentamycin and streptomycin [33]. The genome of E. faecalis CLB21560 also demonstrated a similar combination of genes, hence confirming the AME phenotype of this strain.
Vancomycin resistant E. faecalis (VREF) has emerged as major pathogen in nosocomial infections. Rengaraj R et al., reported highest prevalence of van A and B genes in VREF isolated from clinical specimens [34]. The vancomycin resistant phenotype is aided by the presence of a cluster of genes encoding cell wall modification precursors that exhibits poor affinity towards vancomycin [35]. Here, the normal peptidoglycan precursor with D-alanyl-D-alanine is replaced with D-alanyl-D-lactate, which binds to vancomycin with a lower affinity of 0.001 times when compared to the normal precursors. Several van proteins act synergistically to establish the phenotype [36]. A recent study testing efficacy of genetically engineered bacteriophage φEf11/φFL1C(Δ36)PnisA in treatment of vancomycin resistant strain proved to be a success with vancomycinR strain (E. faecalis V583) exhibiting 99% susceptibility in comparison to 18% in sensitive strains (E. faecalis JH2-2) [37]. The VRE phenotype was observed in two strains analysed in the present study. Additionally, these phenotypes were found to co-occur along with MLSR encoding genes and found in plasmid bearing strains.
The fex(A) gene encodes chloramphenicol resistance which was first identified on the transposon Tn558 in Staphylococcus lentus plasmid. This gene is carried on non-conjugative plasmids in E. faecalis. The gene encodes a transmembrane efflux proteincontaining 475 amino acids forming fourteen transmembrane domains [38,39]. Resistance to trimethoprim-sulfamethoxazole is mediated by dfrG, which encodes the enzyme trimethoprim insensitive dihydrofolate reductase [40]. The dfr genes are mostly found in the plasmids and only a few transmissible dfr genes have been marked in Gram positive organisms, dfrG is one among them and is reported to be detected in Streptococcus pyogenes [41]. The present study also reports the presence of dfrG gene in a strain E. faecalis CLB21560 harboring plasmids which substantiates earlier reports. The prevalence of this gene in E. faecalis was also found to be low (n=1; 6.25%). The present study reiterates the fact that acquired drug resistance should not be ignored, as pathogens assimilating novel genes from the environment might complicate the treatment options and recovery in susceptible patients.
Limitation
Although the present study has certain limitations such as: a) lack of information about the antimicrobial drug resistance exhibited by strains in vitro; b) study restricted towards intrinsic and acquired resistance pertaining to gene clusters excluding point mutations/substitutions; and c) antimicrobial resistance of clinical isolates of E. faecalis from different geographical locations. The present data provides a clear insight about the panel of genes to be detected primarily in case of a treatment failure in suspected E. faecalis infections. A better therapeutic strategy may be adopted for combating such refractory strains.
Conclusion
The trend of antimicrobial drug resistance is creeping into opportunistic and commensal organisms which may in future turn into resistant pathogens making treatments refractory or futile. Hence, meticulous efforts are to be employed to identify, isolate and treat diseases involving resistant species. Alternate treatment modalities such as usage of combinatorial drugs, phage therapy, and probiotics can reduce the incidence of resistant forms. One of the interesting results derived from the study is that E. faecalis harbours one or more resistance encoding genes and that none of the strains were deprived of resistant genotype. To conclude, a vigilant surveillance and focused treatment will help to reduce the load of resistant gene pools in the oral cavity.
E.faecalis FDA ARGOS_338
MLS: Macrolide; Lincosamide; Streptogramin B
[1]. Anderson AC, Jonas D, Huber I, Karygianni L, Wölber J, Hellwig E, Enterococcus faecalis from food, clinical specimens, and oral sites: Prevalence of Virulence Factors in Association with Biofilm FormationFront Microbiol 2016 11:153410.3389/fmicb.2015.0153426793174 [Google Scholar] [CrossRef] [PubMed]
[2]. Sun J, Sundsfjord A, Song X, Enterococcus faecalis from patients with chronic periodontitis: virulence and antimicrobial resistance traits and determinantsEur J Clin Microbiol Infect Dis 2012 31:267-72.10.1007/s10096-011-1305-z21660501 [Google Scholar] [CrossRef] [PubMed]
[3]. Chomicz L, Szubinska D, Piekarczyk J, Wojtowicz A, Piekarczyk B, Starosciak B, Occurrence of subclinical infections of the oral cavity in the insulin treated diabeticsWiad Parazytol 2004 50(2):177-80. [Google Scholar]
[4]. Komiyama EY, Lepesqueur LS, Yassuda CG, Samaranayake LP, Parahitiyawa NB, Balducci I, Enterococcus species in the oral cavity: prevalence, virulence factors and antimicrobial susceptibilityPLoS One 2016 11:e016300110.1371/journal.pone.016300127631785 [Google Scholar] [CrossRef] [PubMed]
[5]. Sedgley C, Molander A, Flannagan SE, Nagel AC, Appelbe OK, Clewell DB, Virulence, phenotype and genotype characteristics of endodontic Enterococcus sppOral Microbiol Immunol 2005 20:10-19.10.1111/j.1399-302X.2004.00180.x15612939 [Google Scholar] [CrossRef] [PubMed]
[6]. Zhu X, Wang Q, Zhang C, Cheung GS, Shen Y, Prevalence, phenotype, and genotype of Enterococcus faecalis isolated from saliva and root canals in patients with persistent apical periodontitisJ Endod 2010 36:1950-55.10.1016/j.joen.2010.08.05321092811 [Google Scholar] [CrossRef] [PubMed]
[7]. Pinheiro ET, Gomes BPFA, Ferraz CCR, Teixeira FB, Zaia AA, Souza-Filho FJ, Evaluation of root canal microorganisms isolated from teeth with endodontic failure and their antimicrobial susceptibilityOral Microbiol Immunol 2003 18:100-03.10.1034/j.1399-302X.2003.00058.x12654099 [Google Scholar] [CrossRef] [PubMed]
[8]. Duggan JM, Sedgley CM, Biofilm formation of oral and endodontic Enterococcus faecalisJ Endod 2007 33:815-18./10.1016/j.joen.2007.02.01617804318 [Google Scholar] [CrossRef] [PubMed]
[9]. Sassone L, Fidel R, Figueiredo L, Fidel S, Faveri M, Feres M, Evaluation of the microbiota of primary endodontic infections using checkerboard DNA–DNA hybridizationOral Microbiol Immunol 2007 22:390-97.10.1111/j.1399-302X.2007.00376.x17949342 [Google Scholar] [CrossRef] [PubMed]
[10]. Pinheiro ET, Gomes BP, Drucker DB, Zaia AA, Ferraz CC, Souza-Filho FJ, Antimicrobial susceptibility of Enterococcus faecalis isolated from canals of root filled teeth with periapical lesionsInt Endod J 2004 37:756-63.10.1111/j.1365-2591.2004.00865.x15479258 [Google Scholar] [CrossRef] [PubMed]
[11]. Reynaud afGeijersstam AH, Ellington MJ, Warner M, Woodford N, Haapasalo M, Antimicrobial susceptibility and molecular analysis of Enterococcus faecalis originating from endodontic infections in Finland and LithuaniaOral Microbiol Immunol 2006 21:164-68.10.1111/j.1399-302X.2006.00271.x16626373 [Google Scholar] [CrossRef] [PubMed]
[12]. Jungermann GB, Burns K, Nandakumar R, Tolba M, Venezia RA, Fouad AF, Antibiotic resistance in primary and persistent endodontic infectionsJ Endod 2011 37:1337-44.10.1016/j.joen.2011.06.02821924178 [Google Scholar] [CrossRef] [PubMed]
[13]. Flannagan SE, Clewell DB, Sedgley CM, A retrocidal plasmid in Enterococcus faecalis: passage and protectionPlasmid 2008 59:217-30.10.1016/j.plasmid.2008.01.00218295881 [Google Scholar] [CrossRef] [PubMed]
[14]. https://www.ncbi.nlm.nih.gov/genome/?term=Enterococcus+faecalis [Google Scholar]
[15]. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Multilocus sequence typing of total genome sequenced bacteriaJ Clin Micobiol 2012 50:1355-61.10.1128/JCM.06094-1122238442 [Google Scholar] [CrossRef] [PubMed]
[16]. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Identification of acquired antimicrobial resistance genesJ Antimicrob Chemother 2012 67:2640-44.10.1093/jac/dks26122782487 [Google Scholar] [CrossRef] [PubMed]
[17]. Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Plasmid Finder and pMLST: in silico detection and typing of plasmidsAntimicrob Agents Chemother 2014 58:3895-903.10.1128/AAC.02412-1424777092 [Google Scholar] [CrossRef] [PubMed]
[18]. Robert E, MUSCLE: multiple sequence alignment with high accuracy and high throughputNucleic Acids Research 2004 32:1792-97.10.1093/nar/gkh34015034147 [Google Scholar] [CrossRef] [PubMed]
[19]. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotesBioinformatics 2010 26:1608-15.10.1093/bioinformatics/btq24920472543 [Google Scholar] [CrossRef] [PubMed]
[20]. Zuckerkandl E, Pauling L, Evolutionary divergence and convergence in proteins, in Evolving Genes and Proteins, edited by V. Bryson and H.J. Vogel 1965 New YorkAcademic Press:97-166.10.1016/B978-1-4832-2734-4.50017-6 [Google Scholar] [CrossRef]
[21]. Kumar S, Stecher G, Tamura K, MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasetsMol Biol Evol 2016 33:1870-74.10.1093/molbev/msw05427004904 [Google Scholar] [CrossRef] [PubMed]
[22]. Felsenstein J, Confidence Limits on phylogenies: An approach using the bootstrapEvolution 1985 39:783-91.10.1111/j.1558-5646.1985.tb00420.x28561359 [Google Scholar] [CrossRef] [PubMed]
[23]. Harbarth S, Kahlmeter G, Kluytmans J, Mendelson M, Hospital GS, Burkert F et al. Implementation of the global action plan on antimicrobial resistance. 2017;32:1-4 [Google Scholar]
[24]. Bozdogan B, Leclercq R, Effects of genes encoding resistance to streptogramins A and B on the activity of quinupristin-dalfopristin against Enterococcus faeciumAntimicrob Agents Chemother 1999 43:2720-25.10.1128/AAC.43.11.272010543753 [Google Scholar] [CrossRef] [PubMed]
[25]. Singh KV, Weinstock GM, Murray BE, An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristinAntimicrob Agents Chemother 2002 46:1845-50.10.1128/AAC.46.6.1845-1850.200212019099 [Google Scholar] [CrossRef] [PubMed]
[26]. Portillo A, Ruiz-Larrea F, Zarazaga M, Alonso A, Martinez JL, Torres C, Macrolide resistance genes in Enterococcus sppAntimicrob Agents Chemother 2000 44:967-71.10.1128/AAC.44.4.967-971.200010722498 [Google Scholar] [CrossRef] [PubMed]
[27]. Hollenbeck BL, Rice LB, Intrinsic and acquired resistance mechanisms in EnterococcusVirulence 2012 15:421-33.10.4161/viru.2128223076243 [Google Scholar] [CrossRef] [PubMed]
[28]. Quiñones Pérez D, Abreu Capote M, Marrero D, Alvarez AB, Ortiz C, Salomé F, Antimicrobial susceptibility and genetic bases for resistance of infection-causing Enterococcus strains in CubaRev Panam Salud Publica 2011 30:549-54.10.1590/S1020-4989201100120000922358401 [Google Scholar] [CrossRef] [PubMed]
[29]. Rôças IN, Siqueira JF Jr, Detection of antibiotic resistance genes in samples from acute and chronic endodontic infections and after treatmentArch Oral Biol 2013 58:1123-28.10.1016/j.archoralbio.2013.03.01023591127 [Google Scholar] [CrossRef] [PubMed]
[30]. Thomas ER, Diane F, Joel EM, John ED, Arie JW, Antibiotic susceptibility of periodontal Enterococcus faecalisJ Periodontol 2013 84:1026-33.10.1902/jop.2012.12005023106507 [Google Scholar] [CrossRef] [PubMed]
[31]. Rathnayake IU, Hargreaves M, Huygens F, Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolatesSyst Appl Microbiol 2012 35:326-33.10.1016/j.syapm.2012.05.00422742879 [Google Scholar] [CrossRef] [PubMed]
[32]. Watanabe S, Kobayashi N, Quiñones D, Nagashima S, Uehara N, Watanabe N, Genetic diversity of enterococci harboring the high-level gentamycin resistance gene aac(6’)-Ie-aph(2”)-Ia or aph(2”)-Ie in a Japanese hospitalMicrob Drug Resist 2009 15:185-94.10.1089/mdr.2009.091719728776 [Google Scholar] [CrossRef] [PubMed]
[33]. Kobayashi N, Alam M, Nishimoto Y, Urasawa S, Uehara N, Watanabe N, Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus aviumEpidemiol Infect 2001 126:197-204.10.1017/S095026880100527111349969 [Google Scholar] [CrossRef] [PubMed]
[34]. Rengaraj R, Mariappan S, Sekar U, Kamalanadhan A, Detection of vancomycin resistance among Enterococcus faecalis and Staphylococcus aureusJ Clin Diagn Res 2016 10(2):DC04-6.10.7860/JCDR/2016/17552.720127042459 [Google Scholar] [CrossRef] [PubMed]
[35]. Gold HS, Vancomycin-resistant enterococci: mechanisms and clinical observationsClin Infect Dis 2001 33:210-19.10.1086/32181511418881 [Google Scholar] [CrossRef] [PubMed]
[36]. Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT, Molecularbasis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptidepeptidogly can precursor by vancomycin resistance proteins VanH and VanABiochemistry 1991 29:10408-15.10.1021/bi00107a0071931965 [Google Scholar] [CrossRef] [PubMed]
[37]. Tinoco JM, Liss N, Zhang H, Nissan R, Gordon W, Tinoco E, Antibacterial effect of genetically-engineered bacteriophage φEf11/φFL1C(Δ36) PnisA on dentin infected with antibiotic-resistant Enterococcus faecalisArch Oral Biol 2017 82:166-70.10.1016/j.archoralbio.2017.06.00528646693 [Google Scholar] [CrossRef] [PubMed]
[38]. Lang KS, Anderson JM, Schwarz S, Williamson L, Handelsman J, Singer RS, Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil by using functional metagenomicsAppl Environ Microbiol 2010 76:5321-26.10.1128/AEM.00323-1020543056 [Google Scholar] [CrossRef] [PubMed]
[39]. Kehrenberg C, Schwarz S, fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicolAntimicrob Agents Chemother 2004 48:615-18.10.1128/AAC.48.2.615-618.200414742219 [Google Scholar] [CrossRef] [PubMed]
[40]. Woods SE, Lieberman MT, Lebreton F, Trowel E, de la Fuente-Núñez C, Dzink-Fox J, Gilmore MS, Fox JG, Characterization of Multi-Drug Resistant Enterococcusfaecalis Isolated from Cephalic Recording Chambers in Research Macaques (Macacaspp.)PLoS One 2017 12:e016929310.1371/journal.pone.016929328081148 [Google Scholar] [CrossRef] [PubMed]
[41]. Bergmann R, Sagar V, Nitsche-Schmitz DP, Chhatwal GS, First detection of trimethoprim resistance determinant dfrG in Streptococcuspyogenes clinical isolates in IndiaAntimicrob Agents Chemother 2012 56:5424-25.10.1128/AAC.01284-1222890758 [Google Scholar] [CrossRef] [PubMed]