Macrolides such as erythromycin constitute an important group of drugs because of their antimicrobial and immunomodulatory activities [1]. Erythromycin is a clinically important antibiotic which binds to a site in 23S rRNA on the large ribosomal subunit 50S close to the peptidyl transferase, near the entrance to the nascent peptide exit tunnel [2]. The resistance to antimicrobial agents is an increasingly global problem worldwide, especially among nosocomial pathogens. Resistance to macrolides such as erythromycin is prevalent among Gram positive cocci [3,4]. Erythromycin resistance can be due to target-site modification by an rRNA-methylating enzyme or by an efflux system. Target-site modification can be expressed either in a constitutive or inducible manner, resulting in co-resistance to Macrolide-Lincosamide-Streptogramin B (MLSB) antibiotics [5]. However, while bacterial resistance is continuously increasing as a result of antibiotic usage in human and veterinary medicine as well as in farms and in agriculture, the industrial antibiotic pipeline has progressively dried up [6]. On several occasions, bacteria isolated from patients are resistant to most of the antibiotics used in the hospital [7].
Staphylococci have become one of the most common causes of nosocomial infections with ability to develop resistance to antibiotics which were hitherto susceptible [8]. There have been several reported cases of erythromycin resistance in major Gram positive cocci pathogens worldwide; in a teaching hospital in Hatay, Turkey a total of 165 out of 298 isolates were resistant to erythromycin, and contained at least one of the erythromycin resistance genes; ermA, ermB, ermC and msrA [9], while erythromycin resistance in alpha-and beta-haemolytic streptococci was mediated through erm and/or mef genes in Hong Kong [10]. High levels of Gram positive cocci resistant to erythromycin has been reported in phenotypic studies carried out in different parts of Africa; in Johannesburg, South Africa 38% of the isolates were resistant to erythromycin [11], also, in Nigeria two separate phenotypic studies reported S. aureus isolates to be 30.7% intermediate and 21.2% resistant [12] and 28.57% resistant [13].
The mechanisms of resistance of erythromycin genes are well known elsewhere, but previous studies in Nigeria have only focused on phenotypic determination of erythromycin resistance [12,13], hence there is little or no information on the determinants of erythromycin resistance. Identifying the mechanisms of Gram positive cocci resistance to erythromycin will be of immense value in curbing and providing appropriate control measures for Gram positive cocci isolates. Therefore, the determinants of erythromycin resistance in clinical Gram positive cocci from Nigerian patients was investigated.
Materials and Methods
Bacterial Isolates
A total of 50 isolates comprising 25 convenient non-duplicate assemblies of staphylococci which were collected for a period of three months between January and March 2015 in a cross-sectional/observational study and 25 enterococci from culture collection previously isolated from stool samples of apparently healthy members of Osogbo community in Osun State as a commensal flora who never visited hospital in the last three months. The staphylococci were from five different tertiary hospitals: University College Hospital, Ibadan; Obafemi Awolowo University Teaching Hospital Complex, Ile-Ife; Ladoke Akintola University of Technology (LAUTECH) Teaching Hospital, Osogbo; LAUTECH Teaching Hospital, Ogbomoso and Federal Medical Centre, Gusau. They were obtained from sick people in these hospitals from varying clinical sites such as urine, blood culture, wound, eye swab, ear swab, high vaginal swab and semen from inpatients and outpatients with varying diseases ranging from urinary tract infections, septicaemia, otitis media and conjunctivitis. The inclusion criteria include all staphylococci isolates from outpatients and inpatients and enterococci from stool samples of healthy members of the community. Also, non-staphylococci isolates from the hospitals including all isolates from intensive care unit and non-enterococci isolates from stool samples of healthy members were excluded from the study including their duplicate strains. Ethical approval was obtained for the study including informed consent from the participants. These isolates were identified using standard procedures as described in Medical Microbiology manual [14].
Antibiotic Susceptibility Testing
The antibiotic susceptibility patterns of all isolates to a panel of antibiotics namely; mupirocin (200 μg), tetracycline (30 μg), linezolid (30 μg), erythromycin (15 μg), gentamicin (30 μg), teicoplanin (30 μg), clindamycin (2 μg), cefoxitin (30 μg) were determined by the disc diffusion method in Mueller-Hinton agar (discs from Oxoid, UK). Staphylococci and enterococci isolates were considered sensitive (≥23 mm), intermediate (14-22 mm) and resistant (≤13 mm) for erythromycin based on the guidelines of Clinical Laboratory Standards Institute [15]. Erythromycin resistant isolates were selected further for MIC to erythromycin using microdilution method [15]. All runs included the control organism Oxford S. aureus NCTC 6571.
Inducible Clindamycin Resistance
Double disk diffusion (D-test) and microdilution tests were used to determine inducible clindamycin resistance [15]. Inocula of bacteria were prepared to 0.5 McFarland standards and sterile swab stick was used to streak the Mueller-Hinton agar. Erythromycin disc (15 μg) was placed 20 mm apart from a clindamycin disc (2 μg) on the surface of Mueller-Hinton plate and incubated at 37°C for 24 hour.
DNA Extraction and Amplification of Erythromycin Genes
DNA was extracted from 500 μL overnight Mueller-Hinton broth of each isolates of staphylococci and enterococci using lysostaphin and lysozyme to digest the cell wall as previously described by Alli O et al., [16]. Conventional PCR was used to detect genes encoding resistance to erythromycin (ermA, ermB, ermC, mefA, msrA and msrB) using a GeneAmp PCR system 9700 thermal Cycler (Applied Biosystems) [Table/Fig-1] [17,18]. Amplimers resulting from these PCR reactions were sequenced to confirm the identity and specific variant of each gene identified and sequences were aligned to known reference sequences using ClustalW [19].
Primers used for the amplification of erythromycin resistance genes.
| Gene | Primer Sequence | Annealing Temperature (°C) | ProductSize (bp) |
|---|
| ermA | ATGAACCAGAAAAACCCTAAA | 52 | 732 |
| GGCTTAGTGAAACAATTTGTAAC |
| ermB | GGCGGATGAACAAAAATATAAAATA | 55 | 738 |
| GCGTTATTTCCTCCCGTTAAA |
| ermC | GGCATGAACGAGAAAAATATAAA | 46 | 735 |
| GCTAATATTGTTTAAATCGTCAAT |
| mefA | AGTATCATTAATCACTAGTGC | 50 | 367 |
| TTCTTCTGGTACAAAAGTGG |
| msrA | CGATGAAGGAGGATTAAAATG | 50 | 1737 |
| CATGAATAGATTGTCCTGTTAATT |
| msrB | TATGATATCCATAATAATTATCCAATC | 50 | 595 |
| AAGTTATATCATGAATAGATTATCCTATT |
Statistical Analysis
Data were analysed using statistical package within the Epi-info software for Centre for Disease Control and Prevention, USA. Chi-square test was used to determine the association between various resistance genes and erythromycin resistance in S. aureus and CoNS. The p-value <0.05 was considered to be statistically significant.
Results
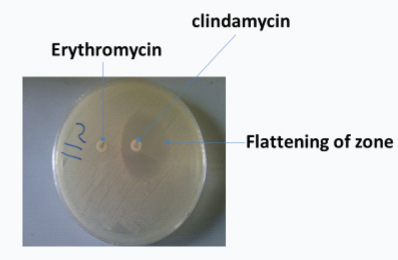
The susceptibility patterns of all the strains against a panel of antimicrobial agents including erythromycin are reported in [Table/Fig-2]. The antimicrobial susceptibility pattern revealed various degree of resistance to the antibiotics. Highest degree of antibiotic resistance to S. aureus was obtained for gentamicin 81.3%, this was followed by cefoxitin 56.3%, while CoNS had highest resistance of 88.9% to cefoxitin while gentamicin and clindamycin 44.4% each. The highest degree of antibiotic resistance to Enterococcus was recorded for tetracycline, 18/25 (72%) while linezolid was 100% sensitive (panel III) [Table/Fig-2]. A flattening of the zone of inhibition around the clindamycin disc proximal to the erythromycin disc was considered as a positive result. Only one of the staphylococcal isolates showed inducible clindamycin resistance [Table/Fig-3].
Antimicrobial disc susceptibility pattern of bacteria isolates.
| Antibiotic (μg/mL) | Resistant (%) | Intermediate (%) | Sensitive (%) |
|---|
| Panel I: 16 Staphylococcus aureus |
| Mupirocin (200) | 1 (6.3) | 6 (37.5) | 9 (56.2) |
| Tetracycline (30) | 8 (50) | 0 (0) | 8 (50.0) |
| Linezolid (30) | 2 (12.5) | 0(0) | 14 (87.5) |
| Erythromycin (15) | 8 (50.0) | 7 (43.8) | 1 (6.2) |
| Gentamicin (30) | 13 (81.2) | 0 (0) | 3 (18.8) |
| Teicoplanin (30) | 1 (6.3) | 2 (12.5) | 13 (81.2) |
| Clindamycin (2) | 8 (50) | 0 (0) | 8 (50.0) |
| Cefoxitin (30) | 9 (56.2) | 0(0) | 7 (43.8) |
| Panel II: 9 CoNS |
| Mupirocin (200) | 0 (0) | 3 (33.3) | 6 (66.7) |
| Tetracycline (30) | 3 (33.3) | 2 (22.2) | 4 (44.4) |
| Linezolid (30) | 1 (11.1) | 0 (0) | 8 (88.9) |
| Erythromycin (15) | 3 (33.3) | 0 (0) | 6 (66.7) |
| Gentamicin (30) | 4 (44.4) | 1 (11.1) | 4 (44.4) |
| Teicoplanin (30) | 0 (0) | 0 (0) | 9 (100) |
| Clindamycin (2) | 4 (44.4) | 1 (11.1) | 4 (44.4) |
| Cefoxitin (30) | 8 (88.9) | 0 (0) | 1 (11.1) |
| Panel III: 25 Enterococci |
| Mupirocin (200) | NA | NA | NA |
| Tetracycline (30) | 18 (72) | 0 (0) | 7 (28) |
| Linezolid (30) | 0 (0) | 0(0) | 25 (100) |
| Erythromycin (15) | 0 (0) | 3 (12) | 22 (88) |
| Gentamicin (30) | NA | NA | NA |
| Teicoplanin (30) | 3 (12) | 0 (0) | 22 (88) |
| Clindamycin (2) | NA | NA | NA |
| Cefoxitin (30) | NA | NA | NA |
NA: Not applicable; CoNS: Coagulase negative staphylococcus
Inducible Clindamycin Resistance test: An inducible clindamycin resistance showing flattening of the zone of inhibition around the clindamycin disc proximal to the erythromycin disc.
The MICs of the two species of staphylococci to erythromycin showed MIC50 and MIC90 of 2 and >64 μg/mL respectively while the MIC of enterococci strains against erythromycin showed MIC50 and MIC90 of 1 and >64 μg/mL respectively. In staphylococci 32% (8/25) had erythromycin resistance of MIC between 8 and >64 μg/mL while 40% (10/25) showed intermediate MIC between 1 and 4 μg/mL with a total of 72% resistance including intermediate. Similarly, enterococci had 28% erythromycin resistance and intermediate each [Table/Fig-4].
Minimum Inhibitory Concentration of 50 Gram positive cocci isolates to erythromycin.
| Organism (No. of Strains) | MIC50 (μg/mL) | MIC90 (μg/mL) | Sensitive(%) | Range (μg/mL) |
|---|
| S. aureus (16) | 2 | >64 | 4 (25.0) | 0.06-64 |
| CoNS (9) | 2 | >64 | 3 (33.3) | 0.06-64 |
| Enterococci (25) | 1 | >64 | 11 (44.0) | 0.06-64 |
CoNS: Coagulase negative staphylococcus
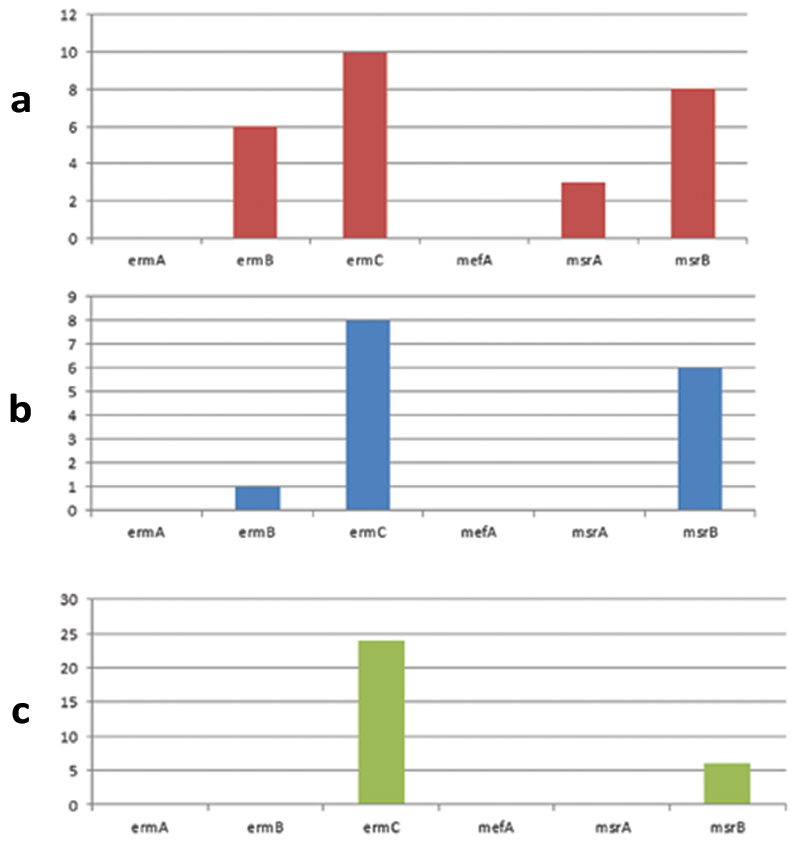
The genotypic properties of the isolates are shown in [Table/Fig-5]. The distribution of erythromycin resistance genes in each of the isolates is shown in [Table/Fig-6]. The ermC was the most prevalent resistant gene in S. aureus, 10/16 (62.5%) followed by msrB, 8/16 (50.0%), msrA was the least prevalent 3/16 (18.8%); these were found in both inpatient and outpatient [Table/Fig-6a]. The ermC and msrB were the most prevalent resistant genes in CoNS; 8/9 (88.9%) and 6/9 (66.7%) respectively, while ermB was the least prevalent 1/9 (11.1%) [Table/Fig-6b]. In enterococci strains, ermC was the most prevalent gene; 23/25 (92.0%) followed by msrB 6/25 (24.0%), while ermA, ermB, msrA and mefA were not found [Table/Fig-6c]. Six strains had ermC and msrB. It is noteworthy that ermA and mefA were consistently not amplified in all the three species and some isolates carried multiple genes of 2 to 4 [Table/Fig-5]. Statistically, there was no association between the presence of ermB gene and erythromycin resistance in S. aureus (χ2=0.33, p=0.56), and same for CoNS (χ2=0.141, p=0.71). Furthermore, no association was found between the presence of ermC and erythromycin resistance in S. aureus (χ2=1.42, p=0.23), neither for CoNS (χ2=0.14, p=0.71).
Genotypic properties of Gram positive cocci isolates.
| Lab No | Specimen | Source | In/Out patient | MIC (μg/mL)Erythromycin | Genes |
|---|
| ermA | ermB | ermC | mefA | msrA | msrB |
|---|
| Staphylococcus aureus |
| S1 | Blood | UCH | Inpatient | 16 | - | - | + | - | - | - |
| S2 | Urine | LTH (Os) | Inpatient | 0.06 | - | + | + | - | + | + |
| S3 | Urine | LTH (Og) | Inpatient | 0.25 | - | - | + | - | - | + |
| S4 | Wound | UCH | Outpatient | 4 | - | - | + | - | - | + |
| S5 | Ear swab | LTH (Os) | Inpatient | 4 | - | - | - | - | - | - |
| S7 | Eye swab | LTH (Og) | Inpatient | 2 | - | + | + | - | + | + |
| S8 | Urine | UCH | Inpatient | 2 | - | + | + | - | - | + |
| S10 | Wound | FMC | Inpatient | 0.5 | - | + | + | - | - | - |
| S11 | Wound | FMC | Inpatient | 8 | - | + | - | - | - | - |
| S12 | Urine | FMC | Outpatient | 8 | - | + | - | - | - | - |
| S13 | Urine | LTH (Og) | Inpatient | >64 | - | - | - | - | - | + |
| S14 | Wound | OAUTHC | Outpatient | 1 | - | - | - | - | - | - |
| S16 | Urine | FMC | Outpatient | 8 | - | - | + | - | + | + |
| S21 | Blood | LTH (Os) | Inpatient | 0.5 | - | - | + | - | - | - |
| S24 | Wound | LTH (Os) | Inpatient | 4 | - | - | - | - | - | - |
| S25 | Semen | FMC | Outpatient | 1 | - | - | + | - | - | + |
| Coagulase Negative Staphylococcus |
| S6 | Urine | LTH (Og) | Inpatient | 0.5 | - | - | - | - | - | - |
| S9 | Blood | FMC | Inpatient | 0.5 | - | - | + | - | - | + |
| S15 | Wound | FMC | Inpatient | >64 | - | - | + | - | - | - |
| S17 | Blood | FMC | Inpatient | 2 | - | - | + | - | - | + |
| S18 | Wound | FMC | Inpatient | 2 | - | - | + | - | - | + |
| S19 | Wound | LTH (Og) | Inpatient | 4 | - | - | + | - | - | - |
| S20 | Ear swab | LTH (Og) | Outpatient | 8 | - | + | + | - | - | + |
| S22 | HVS | FMC | Outpatient | >64 | - | - | + | - | - | + |
| S23 | Urine | FMC | Inpatient | 0.25 | - | - | + | - | - | + |
| Enterococcus spp |
| E1 | Stool | Community | NA | 0.5 | - | - | - | - | - | - |
| E2 | Stool | Community | NA | 64 | - | - | + | - | - | + |
| E3 | Stool | Community | NA | 0.5 | - | - | + | - | - | + |
| E4 | Stool | Community | NA | 1 | - | - | + | - | - | + |
| E5 | Stool | Community | NA | 0.25 | - | - | + | - | - | + |
| E6 | Stool | Community | NA | 4 | - | - | - | - | - | - |
| E7 | Stool | Community | NA | >64 | - | - | + | - | - | + |
| E8 | Stool | Community | NA | 4 | - | - | + | - | - | + |
| E9 | Stool | Community | NA | >64 | - | - | + | - | - | - |
| E10 | Stool | Community | NA | 2 | - | - | + | - | - | - |
| E11 | Stool | Community | NA | 0.25 | - | - | + | - | - | - |
| E12 | Stool | Community | NA | 0.125 | - | - | + | - | - | - |
| E13 | Stool | Community | NA | 0.5 | - | - | + | - | - | - |
| E14 | Stool | Community | NA | 1 | - | - | + | - | - | - |
| E15 | Stool | Community | NA | 0.125 | - | - | + | - | - | - |
| E16 | Stool | Community | NA | 16 | - | - | + | - | - | - |
| E17 | Stool | Community | NA | 4 | - | - | + | - | - | - |
| E18 | Stool | Community | NA | 0.5 | - | - | + | - | - | - |
| E19 | Stool | Community | NA | 0.25 | - | - | + | - | - | - |
| E20 | Stool | Community | NA | 8 | - | - | + | - | - | - |
| E21 | Stool | Community | NA | 8 | - | - | + | - | - | - |
| E22 | Stool | Community | NA | 2 | - | - | + | - | - | - |
| E23 | Stool | Community | NA | 0.5 | - | - | + | - | - | - |
| E24 | Stool | Community | NA | 8 | - | - | + | - | - | - |
| E25 | Stool | Community | NA | 0.25 | - | - | + | - | - | - |
NA: Not Applicable; LTH (Os): Ladoke Akintola University Teaching Hospital; Osogbo; LTH (Og): Ladoke Akintola University Teaching Hospital, Ogbomoso; FMC: Federal Medical Centre; Gusau OAUTHC: Obafemi Awolowo University Teaching Hospital Complex, Ile-Ife; UCH: University College Hospital, Ibadan; Community: healthy community members
Distribution of erythromycin resistance genes in; (a) S. aureus; (b) CoNS; (c) Enterococcus spp.
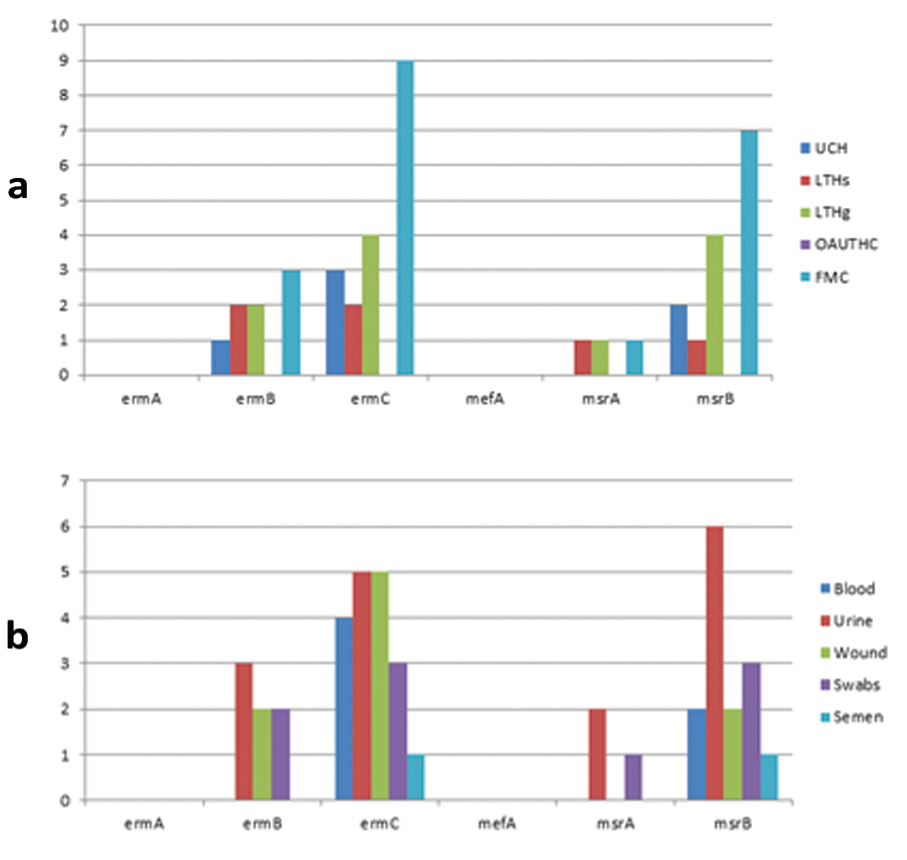
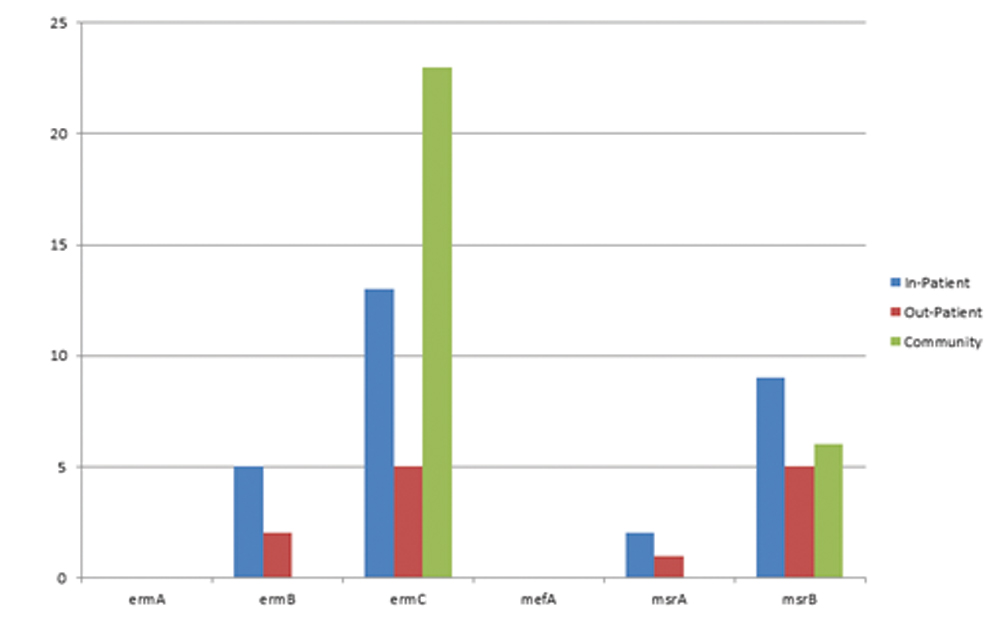
Similarly, the sources of the isolates showed that FMC Gusau had the highest number of genes; ermB, ermC and msrB of 3 (12.0%), 7 (28.0%) and 9 (36.0%) respectively while Ladoke Akintola University of Technology Teaching Hospital Ogbomoso (LTH Ogbomoso) had 1 (4.0%) ermB, while ermC and msrB had 4 (16.0%) each. Isolates from Obafemi Awolowo University Teaching Hospital Complex (OAUTHC) had no resistant gene [Table/Fig-7], Also, inpatients had more frequency of the genes compared to outpatients, while community or carrier isolates which are essentially enterococci had 23/25 (92.0%) ermC, 6 (24.0%) msrB with no other genes present [Table/Fig-8].
Distribution of erythromycin genes according to the sources of the isolates.
Distribution of erythromycin genes amongst the community, inpatients and outpatients.
Discussion
The resistance level of Staphylococcus spp. has been on the increase as a result of widespread utilisation and uncontrolled use of erythromycin [20]. The MIC confirmed the results of disc diffusion that showed high level of erythromycin resistance; S. aureus was more resistant at 75% followed by CoNS 66.7%, all the isolates had MIC90 >64 μg/mL. There is possibility of intermediate susceptibility becoming full blown resistance if clinical use of erythromycin is not controlled. The presence of inducible clindamycin resistance in one of the staphylococci isolates indicates that inducible clindamycin resistance may be a major problem in the future if appropriate measures are not applied. Previous studies have shown a varying level of inducible resistance among the staphylococci [5,21].
Erythromycin resistance can be caused by several mechanisms, the predominant form being target modification mediated by one or more erm genes encoding a 23S rRNA methylase. Target site modification is mediated by the presence of erm genes; ermA, ermB and ermC [22]. The incidence may be influenced by geographical locations including variability from hospital to hospital, the source or origin of isolates, clinical samples or patient types and so on. The ermC was the most frequent gene, found in high number in all the isolates; enterococci had the highest followed by S. aureus and CoNS. Similarly, it was found more in inpatient than outpatient and this applies to other genes, overall more resistant genes were found in inpatients than outpatients. ermC has been regarded as the most widely disseminated and clinically important determinant of MLSB resistance in staphylococci [23]. A study conducted in south western part of Nigeria showed that 4 (two S. epidermidis, one S. haemolyticus and, S. cohnii each) out of five erythromycin resistant isolates possessed ermC genes, while ermA and ermB genes were absent [24]. In other countries such as Italy and Tunisia, ermC was also reported as the prevalent gene in clinical isolates of erythromycin resistant S. epidermidis [25,26]. One of the erythromycin resistant S. haemolyticus strain was found to possess the msrA gene which encoded an ATP-dependent efflux pump conferring resistance to 14- and 15-membered macrolides [7]. ermA was not found in both staphylococci and enterococci isolates, this is a sharp contrast to some previous studies where varying prevalence of ermA were found [27,28] while ermB was not found in enterococci it was found in staphylococci isolates (S. aureus and CoNS).
The resistance to erythromycin caused by the presence of macrolide efflux pumps in staphylococci (encoded by msrA, msrB or mefA) has also been documented [30]. The msrA was present only in S. aureus with a low frequency while msrB was found both in the staphylococci and enterococci isolates and was the most predominant gene after ermC. It was also observed that isolates which possess msrA also have msrB; the genes were found in different geographical locations and in different clinical sites. This study however revealed that carriage of msrA may not be connected with nosocomial acquisition as it was found in both inpatients and outpatients. Previous studies reported the prevalence of msrA to range from none to low [9,30]. The mefA gene was not found in both the staphylococci and enterococci isolates in present study.
All the erythromycin resistant isolates were positive for one or more of ermB, ermC, msrA or msrB. There was no association between the presence of erythromycin resistance genes, ermB; ermC and erythromycin resistance in S. aureus and CoNS. FMC Gusau had the highest frequency of ermB, ermC and msrB. The ermB, ermC, msrA and msrB genes may be the genes responsible for erythromycin resistance as found in this study. In general, major heterogeneity was detected in erythromycin resistance as regards the genotypes and the location of the hospitals. Overall, there is no particular pattern of MIC for the presence of erm or msr variants indicating there may be other mechanisms of resistance. A larger sample to determine the prevalence of these genes collected from different parts of the country may be necessary to establish their circulation in Nigeria and detailed molecular mechanisms will be of immense value to curbing the erythromycin resistance.
Limitation
This present study was limited because there was no grant hence, the use of small sample size and lack of detailed epidemiological study.
Conclusion
In conclusion, high level antibiotic resistance was found in both hospital and community isolates. The ermC and msrB representing ribosomal methylation and efflux respectively are the main resistant determinants with major genotypic heterogeneity in the staphylococci from the hospitals and enterococci from healthy community members.
NA: Not applicable; CoNS: Coagulase negative staphylococcus
CoNS: Coagulase negative staphylococcus
NA: Not Applicable; LTH (Os): Ladoke Akintola University Teaching Hospital; Osogbo; LTH (Og): Ladoke Akintola University Teaching Hospital, Ogbomoso; FMC: Federal Medical Centre; Gusau OAUTHC: Obafemi Awolowo University Teaching Hospital Complex, Ile-Ife; UCH: University College Hospital, Ibadan; Community: healthy community members