Leptospirosis has been considered as a neglected zoonotic disease by World Health Organization and has been classified as an emerging or reemerging infectious disease [1,2]. Leptospirosis is an infectious disease caused by Leptospira interrogans complex which has over 23 serogroups and more than 200 serovars based on the expression of surface exposed lipopolysaccharides. The genus Leptospira contains two species-the pathogenic Leptospira interrogans and the non-pathogenic Leptospira biflexa [3]. Leptospirosis has been reported to be endemic in several parts of South India such as Kerala, Tamil Nadu, Karnataka, Pondicherry and Andamans [4]. Leptospirosis has been a grossly underreported disease in India.
The probable reason might be lack of awareness, lack of clinical suspicion and lack of active surveillance [5]. The diagnosis of leptospirosis is mainly based on serological tests in which the gold standard is Microscopic Agglutination Test (MAT) which is also the most wide spread method. Recently, diagnosis of leptospirosis through PCR based methods has been developed. The real time quantitative Polymerase Chain Reaction (qPCR) have redefined the reference standard as the sensitivity of qPCR exceeds that of culture and MAT and can detect low level of leptospires in the leptospiremic phase, the time when accurate diagnosis is most required [6,7]. In this study use of qPCR in diagnosis of leptospirosis and also its role in prognosis of the disease was analyzed.
Materials and Methods
This study was carried out from April 2013 to April 2016 in a tertiary care center, Mysore, Karnataka, India. Hundred conventional inhouse PCR positive cases for leptospirosis which were positive using serological techniques like MAT and ELISA were included in this study so as to determine the bacterial load. Blood samples were collected from patients suspected of leptospirosis and DNA isolation was done according to the manufacturer’s instruction (HELINI BIOMOLECULES, DNA extraction kit, Chennai). The concentration of isolated DNA was checked using nanodrop spectrophotometer. The isolated DNA was stored in deep freezer at -20°C until use. Ethical clearance was obtained from the institutional ethical committee. Details of patient including the sample number, patient name, age, sex, date of collection, address, duration of illness, and symptoms of the illness were also recorded.
Inclusion Criteria
All patients clinically suspected of having leptospirosis were included in this study during the initial phase and only those samples positive with IgM ELISA, MAT and PCR were selected for this study.
Exclusion Criteria
All patients below 5 years of age were excluded from this study as leptospirosis is considered as an occupational hazard generally. Thirty two cases were excluded according to the exclusion criteria.
The Real-Time PCR kit was procured from Helini Biomolecules, Chennai, India. The procedures were performed according to the manufacturer’s instruction. The primers are based on LipL32 gene of pathogenic Leptospira species.
Negative control set up: A 5 μL of nuclease free water was included instead of purified DNA sample as negative control.
Quantitative positive controls set up: All positive controls prepared from QS1 to QS5 in the place of DNA were included.
Sample collection: Three ml (3 mL) of blood was collected in EDTA tube for patients admitted during the first week of illness, spinned at the 5000 rpm for 5 minutes. The separated plasma on the top was stored at -20°C.
Statistical Analysis
The data was entered in MS-Excel and analysed using SPSS software (version: 22). Tables and diagrams are shown wherever needed. The correlation of bacterial load and various laboratory parameters was done using Spearman’s Rank correlation coefficient.
Results
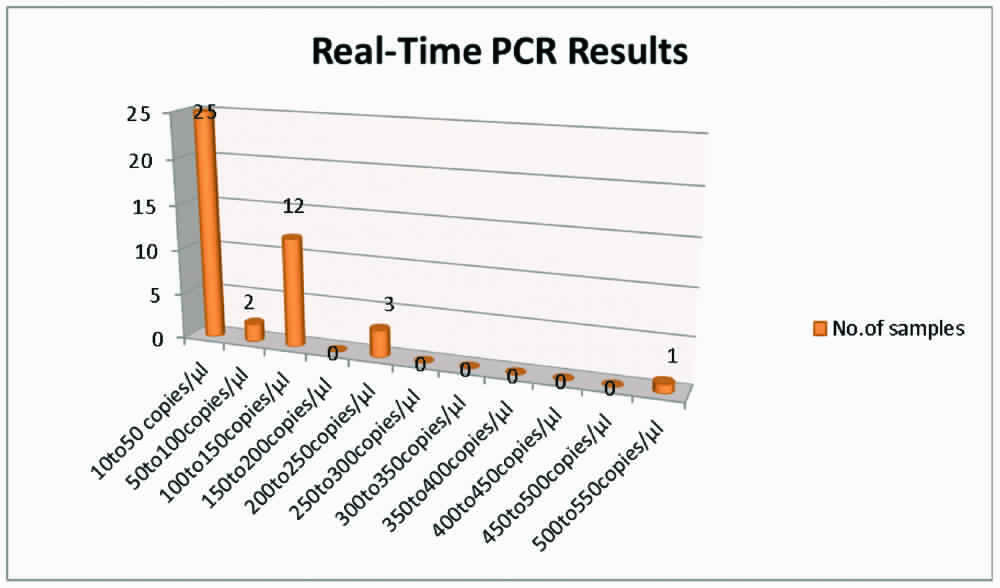
Out of the 100 in-house PCR positive cases, only 43 gave a positive result with Real-Time PCR. A low bacterial load of 10 to 50 copies/μL were seen in 25(58%) cases, 50 to 100 copies/μL in two cases (5%), 100 to 150 copies/μL in 12 cases (28%), 200 to 250 copies/μL in three cases (7%) and 500 to 550 copies/μL in one case (2%). [Table/Fig-1] shows the Real-Time PCR results.
Clinical Profile of Patients Positive with Real-time PCR (N=43)
Thrombocytopenia was seen in 19 of the 43 patients (44%), followed by kidney diseases-13 (30%), hepatomegaly-12 (28%), splenomegaly-nine (21%), pulmonary diseases-eight (19%), meningitis-seven (16%), gastroenteritis-three (7%) and lymphadenitis-two (5%). [Table/Fig-2] shows the bacterial load seen in patients and different complications associated with each bacterial load.
Clinical profile of patients positive with Real-Time PCR.
| Bacterial load (copies/μL) | Complication |
|---|
| 10 (17 cases) | Meningitis,Gastroenteritis (2 cases),Renal involvement+splenomegaly,Renal involvement+splenomegaly+thrombocytopenia,Renal involvement+thrombocytopenia,Hepatic+splenic involvement,Hepatic involvement+thrombocytopenia, (3 cases)Lymphadenitis (2 cases),Pulmonary+hepatic+splenic involvement,Pulmonary involvement,Thrombocytopenia (3 cases) |
| 12 | Pulmonary+renal involvement |
| 15 | Pulmonary involvement |
| 17 (2 cases) | Gastroenteritis,renal involvement |
| 22 | Hepatic+splenic involvement+thrombocytopenia |
| 23 | Renal involvement |
| 24 | Renal+splenic involvement+thrombocytopenia |
| 26 | Pulmonary involvement +thrombocytopenia |
| 56 | Meningitis |
| 58 | Meningitis+hepatic involvement |
| 100 (3 cases) | Kidney injury,Pulmonary involvement,Pulmonary involvement +thrombocytopenia |
| 108 (2 cases) | CKD,Hepatic+splenic involvement |
| 113 (2 cases) | CKD,Renal involvement+thrombocytopenia |
| 115 (3 cases) | CKD+thrombocytopenia,Hepatic+splenic involvement (2 cases) |
| 120 | Pulmonary involvement |
| 125 | Renal involvement+thrombocytopenia |
| 209 | Meningitis+thrombocytopenia |
| 212 (2 cases) | Meningitis+thrombocytopenia+hepatic involvement |
| 515 | Meningitis+thrombocytopenia |
An increased bacterial load was seen associated with meningitis combined with thrombocytopenia and hepatomegaly. Patients with chronic kidney disease also showed high bacterial load. It was difficult to categorize complications based on the bacterial load as majority of the complications fell in the least and the highest bacterial load category.
Laboratory Parameters of Real-Time PCR Positive Cases
Blood Urea Nitrogen (BUN), Aspartate Transaminase (AST), Alanine Transaminase (ALT) along with the potassium levels were seen elevated. BUN was elevated in 21 out of 43 confirmed cases (48.8%), ALT in 28 out of 43 cases (65.1%), AST in 30 out of 43 cases (69.8%) and potassium levels were increased in 20 out of 43 cases (46.5%). Other laboratory parameters like Alkaline phosphatase was seen elevated in 18 out of 43 cases, Sodium levels in 17 out of 43 cases (39.5%), Erythrocytic Sedimentation Rate (ESR) in 16 out of 43 cases (37.2%) and creatinine in 13 out of 43 cases (30.2%). Similarly decrease was also observed in few parameters. Decreased WBC count was seen in 11 of 43 cases (25.6%), total proteins were decreased in 12 out of 43 cases (27.9%), Chloride decrease was seen in 7 out of 43 cases (16.3%) and Sodium levels decreased in 10 out of 43 cases (23.2%). [Table/Fig-3] shows the detailed laboratory profile of Real-Time PCR positive cases.
Laboratory parameters of Real-Time PCR positive patients.
| S. No. | Investigation | Normal Range | Number of Cases (N=43) Elevated | % | N=43 Decreased | % | N=43 Normal | % |
|---|
| 1 | Platelet Count | 1.5 to 4 lacs/cumm | 0 | 0 | 19 | 44.2% | 24 | 55.8% |
| 3 | White Blood cells | 4000 to 11000 cells/cumm | 6 | 14% | 11 | 25.6% | 26 | 60.4% |
| 6 | Creatinine (mg/dL) | 0.7-1.4 mg/dL | 13 | 30.2% | 6 | 14% | 24 | 55.8% |
| 7 | BUN (mg/dL) | 15-40 mg/dL | 21 | 48.8% | 5 | 11.6% | 17 | 39.5% |
| 8 | Alkaline Phosphatase | 37-306 U/L | 18 | 41.9% | 5 | 11.6% | 20 | 46.5% |
| 9 | Alanineamino Transferase/SGPT (IU/L) | Upto 40 U/L | 28 | 65.1% | 0 | 0 | 15 | 34.9% |
| 10 | Aspartate AminoTransferase/SGOT (IU/L) | Upto 37 U/L | 30 | 69.8% | 0 | 0 | 13 | 30.2% |
| 12 | Total Bilirubin | 0.1-1 mg/dL | 2 | 4.7% | 0 | 0 | 41 | 95.3% |
| 13 | Direct Bilirubin | Upto 0.4 mg/L | 1 | 2.3% | 0 | 0 | 42 | 97.7% |
| 14 | Total Proteins | 6-8 gm/dL | 1 | 2.3% | 12 | 27.9% | 30 | 69.8% |
| 15 | Albumin | 3.5-5.2 gm/dL | 1 | 2.3% | 0 | 0 | 42 | 97.7% |
| 16 | A/G | 0.8-2.0 | 2 | 4.7% | 0 | 0 | 41 | 95.3% |
| 17 | ESR | Upto 10 mm in 1 Hr | 16 | 37.2% | 0 | 0 | 27 | 62.8% |
| 18 | Haemoglobin (g/dL) | 12-15 g/dL | 6 | 14% | 1 | 2.3% | 36 | 83.7% |
| 20 | K | 136-145 mEq/L | 20 | 46.5% | 9 | 20.9% | 14 | 32.6% |
| 21 | Na | 3.5-5.5 mEq/L | 17 | 39.5% | 10 | 23.3% | 16 | 37.2% |
| 22 | Cl | 95-110 mEq/L | 4 | 9.3% | 7 | 16.3% | 32 | 74.4% |
The correlation between bacterial load and various parameters were analysed using Spearman’s rank correlation. There was a significant negative correlation between haemoglobin percentage and bacterial load in patients who had kidney involvement {r=-0.601, p-value=0.039}. According to the Spearman’s rank correlation, as the bacterial load increased, haemoglobin level decreased. There was a highly significant correlation noticed between bacterial load and blood urea nitrogen in those patients in whom kidney involvement was seen {r=0.805, p-value=0.016} and also highly significant correlation was found between bacterial load and AST and ALT respectively {r=0.938, p-value=0.001, r=0.805, p=0.016}. Since the sample size tested by Real-Time PCR is very small, further studies are required to establish these parameters as prognostic markers during leptospirosis.
Discussion
The Real-Time PCRs were introduced as techniques that further improved the conventional PCRs by performing faster and reducing the false positive results caused by carry over contaminations (contamination from products from previous reactions) [8,9]. Several quantitative PCR protocols targeting different genes have been developed and are claimed to be specific for pathogenic Leptospira and appropriate for diagnostic purposes [7,10-12]. The Real-Time PCR applied in this study was based on the LipL32 gene present only in pathogenic Leptospira species, but a lower diagnostic positivity rate of (43%) was observed when compared to inhouse PCR. In a study by Agampodi SB et al., a new quantitative PCR assay using pathogenic Leptospira-specific 16S ribosomal RNA (rRNA) gene Taqman primers confirmed 58 of 381 cases [13]. According to the study, quantitative leptospiremia in serum or whole blood samples did not directly correlate with clinical manifestations of outcome in this patient population. In present study, the positivity rate of Real-Time PCR was 43% and could find a significant negative correlation between bacterial load and few laboratory parameters was found. As the bacterial load increased, haemoglobin level decreased. Also, there was a highly significant correlation between bacterial load and blood urea nitrogen in patients with kidney involvement (r=0.805, p-value=0.016) and in those patients with liver disease highly significant correlation was observed between bacterial load and AST and ALT respectively (r=0.938, p-value=0.001, r=0.805, p-value=0.016). But due to very small sample size, further studies with a large study population will be required to establish these parameters as prognostic markers.
The positivity rate of Real-Time PCR used in our study is not in concordance with the study done by Fornazari F et al., where they had reported that quantitative PCR presented the highest sensitivity among several techniques to detect leptospires in tissue samples [14]. A study by Ferreira AS et al., using a new quantitative PCR suggested that though the technique is sensitive and specific, the kind of samples used in their study is not essential for an early diagnosis of leptospirosis [15]. So, it is evident that the type of sample used can affect the outcome.
A Real-Time PCR assay based on LipL32 could be a useful tool in the rapid diagnosis of acute leptospirosis, especially in cases with rapid mortality before serology or culture is able to aid in the diagnosis, such as with severe pulmonary haemorrhagic syndrome [16]. Whole blood spiked with 10 leptospires/mL was not determined to be culture positive until 6 weeks after inoculation, but were able to detect the bacteria using the LipL32 Real-Time PCR in another study by Stoddard RA et al., [7]. The higher performance of PCR compared with other current diagnostic techniques was also demonstrated by the fact that they were able to detect leptospires from the sera of patients who died before seroconversion. Thaipadunpanit J et al., in his study compared the diagnostic specificity and sensitivity of two Real-Time PCR assays targeting rrs and LipL32 for diagnosis of leptospirosis in Thailand and reached a conclusion that the use of Real-Time PCR could improve the management of patients attending the hospital within the first few days of the onset of symptoms of leptospirosis [17]. Very few studies have been carried out to identify the prognostic markers depending on the bacterial load. So, such a study in a large scale population might be a starting point in exploring the possibilities of establishing a prognostic marker in the diagnosis of leptospirosis.
However, the basis for a negative Real-Time PCR result but positive inhouse PCR need to be evaluated. The samples subjected to Real-Time PCR were positive with the Gold standard technique MAT and also IgM ELISA. One of the possible explanations for a negative Real –Time PCR is that a very low count in the initial sample associated with a stochastic effect in which the bacterial DNA was present in the aliquot taken for inhouse PCR but not for Real-Time PCR. Prolonged storage of DNA might be another possible explanation that leads to a very poor sensitivity of Real-Time PCR. Though the commercial kit used for this study targeted LipL32 gene specific for pathogenic Leptospira, the lack of sensitivity and precision might have led to a lower detection limit. Further studies with a larger sample size and immediate processing will be able to throw light into all the unanswered questions.
Conclusion
This study was done to analyse the use of quantitative Real-Time Polymerase Chain Reaction (qPCR) in diagnosis of leptospirosis and its role in prognosis of the disease. Out of 100 samples which were positive using conventional PCR, only 43 gave a positive result with Real-Time PCR. The lowest bacterial count obtained through qPCR was 10 copies/μL and highest was 515 copies/μL. Thrombocytopenia was associated with the least and the highest bacterial load whereas an increased bacterial load was seen associated with meningitis combined with thrombocytopenia and hepatomegaly and chronic kidney disease. There was a highly significant correlation noticed between bacterial load and blood urea nitrogen in those patients in whom kidney involvement was seen and also highly significant correlation was found between bacterial load and AST and ALT respectively. Since the sample size tested by Real-Time PCR is very small, further studies are required to establish these parameters as prognostic markers during leptospirosis.