Introduction
Breast cancer, the most complex multifactorial disease, one of the most common malignancies seen in females over the world. Single-Nucleotide Polymorphisms (SNPs) that occur in DNA repair genes are the contributors to cancer development as they lead to alteration in protein function, impair DNA damage responses, and result in loss of efficiency of DNA repair pathways.
Aim
To determine the association of the two polymorphisms (rs25487 and rs1799782) of the gene XRCC1 involved in the Base Excision Repair (BER) pathway with breast cancer risk.
Materials and Methods
In the present study, 200 breast cancer cases and 200 healthy age-matched controls were analysed with regard to the genotype distribution of XRCC1rs1799782 and rs25487 polymorphism using Taqman allelic discrimination assay by Real time PCR. Chi-square test was the statistical method used, and compliance of the genotype frequencies with Hardy-Weinberg equilibrium was verified. The relative risk was assessed by determining the Odds Ratios (OR) and 95% Confidence Intervals (CI). Insilico studies were carried out for identifying the possible functional and deleterious nsSNPs in XRCC1 gene.
Results
With regard to SNPs involved in the genes that play a role in the BER pathway, a significant association was found for the G/A heterozygous genotype of the XRCC1 (rs25487) (OR; 1.79; 95% CI, 1.17-2.73; p-value: 0.006) with the risk of breast carcinoma. Also, an association was observed between the A/A homozygous genotype of the XRCC1 (rs25487) (OR; 2.08; 95% CI, 1.08-4; p-value 0.02) and breast carcinoma risk. There was a lack of association of the CT/TT genotype of the XRCC1 (rs1799782) (OR, 1.12; 95% CI, 0.69-1.80; P 0.63) with breast carcinoma risk. The insilco studies revealed structural variation in XRCC1 gene with respect to rs25487.
Conclusion
The variant rs25487 of XRCC1 gene was associated with the risk of breast carcinoma, but no association was found with regard to frequency of the rs1799782 variant in the XRCC1 gene. The stability prediction and pathogenicity analysis with computational tools revealed the nsSNPrs25487 interfering with function and structure of the XRCC1 protein. The data suggest that the variant of XRCC1 (rs25487) may contribute to breast cancer susceptibility, extensive study linking DNA repair, environmental factors and ethnicity are needed.
Introduction
Worldwide, breast cancer research is a challenging one due to various reasons. The heterogeneous nature of breast cancer, is further complicated because of the differences in histopathological characteristics, stage of the cancer, and molecular subtypes [1-4]. Despite the fact of available treatment modalities, the mortality and incidence of breast cancer is on the rapid rise [5]. The DNA damage that is caused by exogenous and endogenous mutagenic substances is one of the reasons for the disease. If this damage progresses unrepaired, cell death may ensue on one hand, and on the other dysregulated cell growth can be a consequence, which would probably lead to cancer.
The body has several repair mechanisms in place, to repair the damaged DNA and among these the base excision repair pathway which is involved in mending the breaks in the DNA that are caused by oxidation damage [6]. XRCC1 is a gene that plays a vital role in this pathway by interacting with the DNA pol-β polynucleotide kinase enzyme, DNA ligase IIIα, and PARP1, DNA repair machinery enzymes, to fulfill its role [7,8]. The SNP rs25487 of XRCC1 is a functional polymorphism that is characterised by a base change of G to A, which leads to the substitution of glutamine for arginine at exon 10 codon 399 [9,10]. This SNP is present in the polymerase binding domain (poly ADP-ribose) of XRCC1, which is in the conserved region. The SNP rs1799782 is the conserved region that acts as a linker (hydrophobic) between the polymerase-interacting domains and polymerase β domain; thus, the substitution of tryptophan for arginine will lead to a change in the mechanism of interaction with the proteins that are involved in repair of DNA damages [11]. DNA repair processes are an essential part in a cell that helps to maintain genomic stability. A reduction in the repair capacity could lead to accumulation of damages, further mutations, and the development of diseases such as cancer [12]. Therefore, understanding the mechanisms of mutation and polymorphism, either somatic or inherited, of the DNA repair pathway may possibly lead to the development of new antitumour agents and chemopreventive targets, thus making this a critical area of research [12, 13]. The current study aimed to determine the risk attributed by the two polymorphisms in the XRCC1 gene (rs25487 and rs1799782) polymorphisms which plays a major role in base excision repair pathway for breast cancer and to determine the structural and protein modification of the variant XRCC1 by insilico tools.
Materials and Methods
Study Design
The present case control study involved 200 patients with histologically confirmed breast cancer from Southern India. The patients were recruited from the Department of Oncology, Sri Ramachandra Medical College and Research Institute, Chennai, India, from January 2013 to December 2017. The DNA was isolated from the collected samples. The quality and quantity of the DNA is checked by agarose gel electrophoresis and nanodrop. Genotyping was determined by Real time PCR Taqman allelic discrimination assay for rs1799782 and rs25487 of the XRCC1 gene. All statistical analysis was carried by SPSS statistical software ver. 9.0. For the XRCC1 gene computational tools are used to predict the protein stability for the variant XRCC1 protein.
Study Subjects
For all the patients, relevant clinical and pathological data were obtained while obtaining the informed consent [Table/Fig-1]. For healthy volunteers, inclusion criteria were as follows: lack of a previous diagnosis of benign breast disease of any kind; no history of mastectomy, hysterectomy or oophorectomy; absence of a family history of ovarian, breast, endometrial and/or prostate cancer; and no mental or physical disability. With regard to ethnicity, the patients and controls were well matched, and they were also age matched assessed while obtaining consent for participation. The criteria for breast cancer patients to be included in the study were as follows: no previous treatment for cancer and confirmation of breast malignancy with histological diagnosis. Informed consent was obtained from all the subjects involved in the study. Ethical Clearance was obtained from the institutional ethical committee of Sri Ramachandra Medical College and Research Institute (Deemed to be University) (Ref No: IEC-NI/13/APR/33/28).
Demographic features of breast cancer patients in the study.
| Variables | Breast cancer patients (Number/percentage) |
|---|
| Menopausal status |
| Premenopausal | 86 |
| Post Menopausal | 114 |
| Familial/Sporadic |
| Sporadic | 171 |
| Familial | 29 |
| Number of Births |
| <2 births | 151 |
| >2 births | 49 |
| Parous/Nulliparous |
| Parous | 178 |
| Nulliparous | 22 |
| Stage of Cancer |
| Stage I | 16 |
| Stage II | 60 |
| Stage III | 96 |
| Stage IV | 28 |
| Receptor Status |
| ER+ve | 46% |
| PR+ve | 42% |
| Her 2 +ve | 34% |
| Triple Negative | 20% |
Molecular Analysis by Taqman Allelic Discrimination Assay
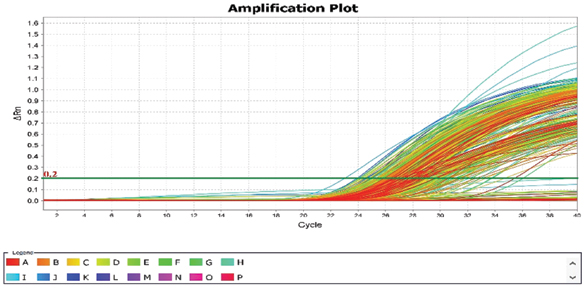
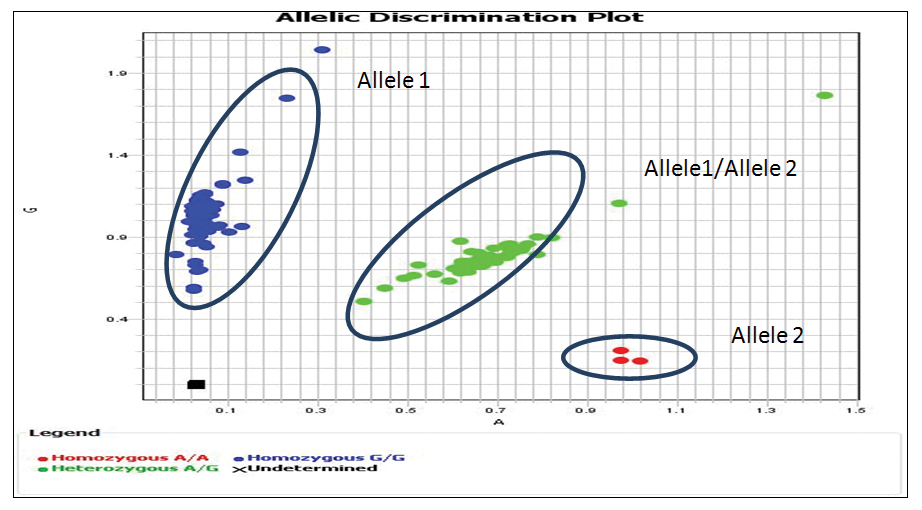
Peripheral blood lymphocyte samples about 3 mL was obtained by veinpuncture and collected into K2-EDTA vacutainers. Genotyping of the SNPs was carried out with real-time polymerase chain reaction technology (Taqman SNP Genotyping Assay, Applied Biosystems, Carlsbad, USA). The isolated DNA was amplified using Taq Gold Polymerase of Taqman PCR master mix in ABI machine Quant studio 6 Flex using sequence specific primers [Table/Fig-2]. The reaction volume was set to 5 μL, consisting of 2.50 μL of (2X) Taqman genotyping master mix, 0.25 μL of (20X) Taqman mutation assay mix and 2.25 μL of the genomic DNA of 10 ng concentration obtained on diluting the DNA with distilled water. Thermal cycle reaction condition is programmed to initial denaturation temperature and timing set at 95°C for 10 minutes followed by further denaturation at temperature set at 95°C for 15 seconds for 40 cycles followed by annealing and extension temperature and timing set at 60°C for one minute in 384 wells in Quant studio 6 Flex machine. Fluorescent signals are obtained for the amplification of each allele by the Taqman probes [Table/Fig-3]. After the amplification by PCR, an end plate read will be performed in ABI machine. The fluorescent measurement taken during the plate read will be used by the sequence detection system and fluorescence the (Rn) values are plotted with the signals obtained from each well [Table/Fig-4,5]. Each fluorescent detector will be a perfect match for the allele 1 and the other for allele 2. The VIC and FAM dye are used for labeling the primers [Table/Fig-2].
Primer Sequences for Taqman allelic discrimination assay real time PCR XRCC1 gene regions.
| rsid | Primer Sequence |
|---|
| rs1799782 | [VIC/FAM]:TCACCTGGGGATGTCTTGTTGATCC[A/G]GCTGAAGAAGAGAGCCCCCGGCCTC |
| rs25487 | [VIC/FAM]:GGGTTGGCGTGTGAGGCCTTACCTC[C/T]GGGAGGGCAGCCGCCGACGCATGCG |
rsid: Reference SNP cluster ID
Representative amplification plot of XRCC1 rs1799782 for the by Taqman allelic discrimination assay by real time PCR.
Representation of genotype detection by end point fluorescence using labelled primers.
The scatter plot is depicted. Taqman allelic discrimination assay by real time PCR Allele1- labelled with VIC and allele 2- labelled with FAM
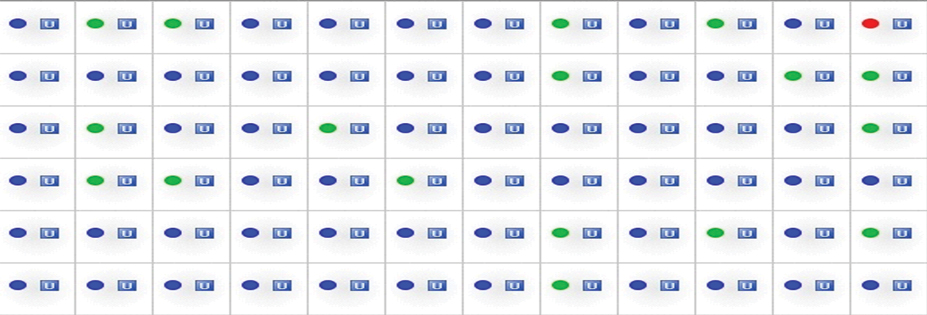
Result grid in 384 well late for Taqman allelic discrimination assay real time PCR representation of rs1799782 polymorphism of XRCC1 gene.
Taqman allelic discrimination assay by real time PCR Allele1- labelled with VIC and allele 2-labelled with FAM
Statistical Analysis
The SPSS statistical software version 9.0 was used for statistical analysis. Chi-square goodness-of-fit test carried out for comparison of expected and observed genotype frequencies for performing Hardy Weinberg equilibrium. Chi-square test was done for comparing the frequencies of the genotype of the polymorphisms between the controls and subjects with breast cancer. Using the genotypes of the wild type as the reference groups the OR and 95% CI was determined. p<0.05 level was considered to be significant.
Prediction of deleterious variants: The protein sequence for XRCC1 gene and its SNP (rs25487) and (rs1799782) was obtained from SWISS-Protein database and from National Centre for Biotechnology Information (NCBI) database respectively. The 3D structure of XRCC1was obtained from PDB database. Five computational tools (I-Mutant Suite, iStable, PolyPhen2, SNAP, and PROVEAN) were used for insilico predictive analysis on the implication of the mutation on the function and structure of the variant XRCC1 protein [Table/Fig-6]. The I-Mutant Suite predicts the protein the variant based on the stability of the protein in to three classes on the basis of the DDG value as neutral stability, large decrease in stability and large increase in stability. SVM based is table integrates the result obtained from the five-prediction analysis with the information of the sequence to analyse the stability of the protein. PolyPhen2 classifies the variant in to three types as benign, probably damaging and possibly damaging. SNAP predicts the variant effect on the function of the protein as neutral and non-neutral based on structural and evolutionary sequence information. The PROVEAN predicts the impact of the variation in the protein sequence due to amino acid substitution as neutral or deleterious [14-18].
Deleterious effects prediction of rs25487 using in silico tools and (XRCC1; and rs1799782 using in silico tools. (XRCC1; 580C>T; Arg194Trp.
| Deleterious effects prediction of rs25487 using in silico tools |
|---|
| Prediction tool | Interpretation |
| I-MUTANT 3 | Decrease (-0.94) |
| I-STABLE | Decrease (0.774155) |
| PolyPhen 2 | Probably damaging (0.979) |
| SNAP 2 | Non-neutral |
| PROVEAN | Deleterious (-3.655) |
| Deleterious effects prediction of rs1799782 using in silico tools |
| Prediction tool | Interpretation |
| I-MUTANT 3 | Decrease (-0.31) |
| I-STABLE | Decrease (0.674266) |
| PolyPhen 2 | Possibly damaging (0.899) |
| SNAP 2 | Non-neutral |
| PROVEAN | Deleterious (-4.418) |
Five computational tools (I-Mutant Suite, iStable, PolyPhen2, SNAP, and PROVEAN) were used for insilico predictive analysis on the implication of the mutation on the function and structure of the variant XRCC1 protein
Results
Hardy-Weinberg Equilibrium
The frequency distribution of the genotype of the polymorphism in the control and of the breast cancer patients was found to be consistent with Hardy Weinberg equilibrium [Table/Fig-7,8].
Allele and genotype frequency distribution of XRCC1 (rs1799782) in Breast cancer affected and control groups.
| Gene | Genotype | Control | Case | OR (95% CI) | p-value |
|---|
| XRCC1 | CC | 158 (79%) | 154 (77%) | Ref | |
| CT | 39 (19.50%) | 43 (21.50%) | 1.13 (0.69-1.84) | 0.62 |
| TT | 3 (1.50%) | 3 (1.500%) | 1.03 (0.20-5.16) | 0.97 |
| CT+TT | 42 (21) | 46 (23) | 1.12 (0.69-1.80) | 0.63 |
| C | 355 (88.75) | 351 (87.75) | Ref | |
| T | 45 (11.25) | 49 (12.25) | 1.10 (0.71-1.69) | 0.66 |
| MAF | 0.11 | 0.12 | | |
| HWP | 0.81 | 0.99 | | |
Association between breast cancer cases and SNP was evaluated by χ2 test. HW compliance was verified for the genotypes. The relative risk was accessed by calculating the odds ratio(OR) and 95% confidence Interval (CI), p<0.05 level was considered to be significant
Allele and genotype frequency distribution of XRCC1 (rs25487) in Breast cancer affected and control groups.
| Gene | Genotype | Control | Case | OR (95% CI) | p-value |
|---|
| XRCC1 | GG | 103 (51.5%) | 73 (36.5%) | Ref | |
| GA | 78 (39%) | 99 (49.5%) | 1.79 (1.17-2.73) | 0.006** |
| AA | 19 (9.5%) | 28 (14%) | 2.08 (1.08-4.00) | 0.02* |
| GA+AA | 97 (48.5) | 127 (63.5) | 1.85 (1.24-2.75) | 0.002** |
| G | 284 (71) | 245 (61.25) | Ref | |
| A | 116 (29) | 155 (38.75) | 1.55 (1.15-2.08) | 0.003** |
| MAF | 0.29 | 0.39 | | |
| HWP | 0.45 | 0.54 | | |
Association between breast cancer cases and SNP was evaluated by χ2 test. HW compliance was verified for the genotypes. The relative risk was accessed by calculating the odds ratio(OR) and 95% confidence Interval (CI), p<0.05 level was considered to be significant; p<0.05 level was considered to be significant, *p<0.05, **p<0.01
Genotyping and Breast Cancer Risk
The genotype frequency of distributions of the XRCC1 rs1799782 and rs25487 polymorphisms between cases affected with breast cancer and healthy controls shown in the [Table/Fig-7,8]. For XRCC1rs1799782 C to T polymorphism, the frequency of homozygous wild type CC was 79% in controls and 77% in cases, heretozygous variant CT was 19.5% and 21.50% between controls and cases and Homozygous mutant TT was 3% in the control and cases. Thus, the distribution frequency is similar in cases and control groups for rs1799782. For rs25487 of XRCC1 gene the homozygous GG; found to be 51.5% and 36.5% in controls and cases. The heterozygous GA; 39% and 49.5% in controls and cases with OR of 1.79 p=0.006. The homozygous AA genotype was 9.5% in controls and 14% in cases with OR 2.08 and p-value=0.02. A significant increase of GA and AA alleles of the gene XRCC1rs25487 was found in patients in comparison with that of controls. The polymorphism rs25487 and rs1799782 the genotype frequencies were compared with clinical parameters and tumor grade [Table/Fig-9,10]. For rs25487 of XRCC1 gene the heterozygous genotype GA was found to be associated with premenopausal females odds ratio (OR) of 2.3, p-value=0.01. There was no association found for rs1799782 with regard to tumour grade and menopausal status [Table/Fig-9,10].
XRCC1 (rs1799782) and (rs25487) genotype among breast cancer patients and control stratified by age at diagnosis.
| XRCC1 (rs1799782) genotype among breast cancer patients and control stratified by age at diagnosis |
|---|
| Genotype (rs1799782) | Controls (%) | Patients (%) | OR | 95% CI | p-value |
|---|
| Premenopausal | N=86 | N=86 | | | |
| CC | 68 (79%) | 70 (81.3%) | | | |
| CT | 17 (19.7%) | 14 (16.2%) | 0.8 | 0.36-1.74 | 0.57 |
| TT | 1 (1.16%) | 2 (2.3%) | 1.94 | 0.17-21.9 | 0.59 |
| Menopausal | N=114 | N=114 | | | |
| CC | 90 (78.9%) | 84 (73.6%) | | | |
| CT | 22 (19.2%) | 29 (25.4%) | 1.41 | 0.75-2.64 | 0.28 |
| TT | 2 (1.75%) | 1 (0.87%) | 0.53 | 0.04- 6.01 | 0.61 |
| XRCC1 (rs25487) genotype among breast cancer patients and control stratified by age at diagnosis |
| Genotype (rs25487) | Controls (%) | Patients (%) | OR | 95% CI | p-value |
| Premenopausal | N=86 | N=86 | | | |
| GG | 52 (60.4%) | 34 (39.5%) | | | |
| GA | 24 (27.9%) | 36 (41.8%) | 2.3 | 1.16-4.49 | 0.015** |
| AA | 10 (11.6%) | 16 (18.6%) | 2.4 | 0.99-6.02 | 0.05 |
| Menopausal | N=114 | N=114 | | | |
| GG | 51 (44.7%) | 39 (34.2%) | | | |
| GA | 54 (47.3%) | 63 (55.2%) | 1.52 | 0.87- 2.65 | 0.13 |
| AA | 9 (7.8%) | 12 (10.5%) | 1.74 | 0.66-4.55 | 0.256 |
Association between premenopausal, menopausal and SNP was evaluated. The relative risk was accessed by calculating the odds ratio(OR) and 95% confidence Interval (CI), p<0.05 level was considered to be significant.
Association between XRCC1 (rs1799782), (rs25487) and tumor grade.
| Association between XRCC1 (rs1799782) and tumor grade |
|---|
| Genotype (rs179982) | Patients % | OR | 95% CI | p-value |
|---|
| Low gradeTumors | High GradeTumors |
|---|
| CC | 55 (72.3%) | 99 (79.8%) | | | |
| CT | 18 (23.6%) | 25 (20.1%) | 0.77 | 0.38-1.5 | 0.46 |
| TT | 3 (96%) | 0 (0) | 3.9 | 0.19-76.9 | 0.37 |
| Association between XRCC1 (rs25487) and tumor grade |
| Genotype (rs25487) | Patients % | OR | 95% CI | p-value |
| Low gradeTumors | High GradeTumors |
| GG | 27 (35.5%) | 46 (37.08%) | | | |
| GA | 34 (44.7%) | 65 (52.4%) | 1.12 | 0.59-2.10 | 0.72 |
| AA | 15 (19.7%) | 13 (10.4%) | 0.50 | 0.21-1.22 | 0.133 |
Association between tumor grade and SNP was evaluated. The relative risk was accessed by calculating the odds ratio (OR) and 95% confidence Interval (CI), p<0.05 level was considered to be significant
In Silico Predictive Functional Analysis
The pathogenicity prediction and stability was analysed using five important tools (I-Mutant Suite, iStable, SNAP, PolyPhen2, and PROVEAN). The analysis showed the variant Arg399Gln to be affecting the structure of the protein as the structure is different from the native protein, affecting the stability of the protein structure [Table/Fig-11,12]. The stability of the structure analysed by I-Mutant suite revealed decrease, largely in the stableness of the variant protein due to the free energy change (DDG value-0.94). The iStable analysis also revealed a decrease in the structure stability due to the variant form. PolyPhen2, analysis by the score of 0.979 revealed the variant Arg399Gln as probably damaging the functional analysis performed through PolyPhen2 classified the mutant (Arg399Gln) as “Probably damaging”, by the score of 0.979. The SNAP and PROVEAN analysis predicted the variant (Arg399Gln) to be deleterious [Table/Fig-6].
XRCC1 399 - GLN (MUTANT).
Variant structure showing Glutamine at positions 399 position
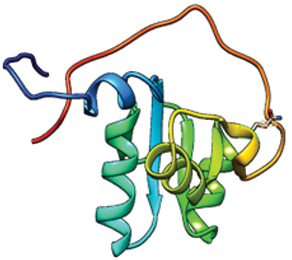
XRCC1 399-ARG (NATIVE).
Native structure showing arginine at positions 399 position.
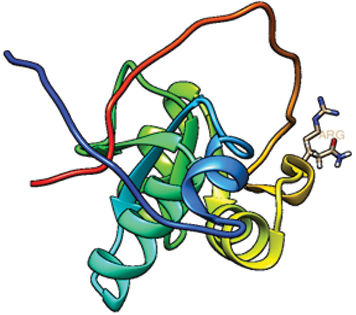
For rs1799782, the stability of the structure analysed by I-Mutant suite revealed decrease largely in the stableness of the variant protein due to the free energy change DDG value -0.31. The iStable analysis also revealed a decrease in the structure stability due to the variant form. PolyPhen2 analysis by the score of 0.899 revealed the variant Arg194Trp as possibly damaging. The SNAP and PROVEAN analysis predicted the variant Arg194Trp to be deletrious [Table/Fig-6].
Discussion
XRCC1 plays a key role as a scaffold protein in the BER pathway and it helps to assemble the protein machineries of the DNA repair complex. XRCC1 has been reported as a candidate gene influencing the risk of cancer. The present study aimed at assessing the association of polymorphisms in XRCC1, rs25487 (Arg399Gln) and rs1799782 (Arg194Trp), with breast cancer risk in a Southern Indian population and found rs25487 to be significantly associated with breast cancer risk. The reactive oxygen species that is generated during normal metabolic processes in the cell and also in response to exogenous genotoxins is one of the main causes of DNA damage [19,20]. The presence of adducts in the DNA has been found in breast ductal epithelial cells and even in tissue of human breast [21-23]. An increased risk of breast cancer is seen with the presence of DNA adducts [24,25]. BER pathway is the one that helps to remove the oxidised and alkylated bases. In addition to XRCC1, PARP1 and OGG1 enzymes are also involved in BER pathway. Polymorphisms in any of these genes may contribute to the risk of developing malignancy.
The XRCC1 polymorphism and its association with other various cancers namely breast cancer, head and neck cancer and lung cancer have been studied [26-28]. The result obtained from the previous studies are not consistent [7,29]. The same is also true for the studies determining the association between XRCC1 polymorphism and breast cancer, where studies have shown conflicting results [10,26,30,31]. Thus, the association between XRCC1 genetic polymorphisms at codons 194 and 399 with regard to the susceptibility to breast cancer is still an open-ended question [32].
The 194Trp codon location is at a special hydrophobic linker region which is the highly conserved region of XRCC1 gene. This region serves as a linker between the polymerase interacting domain (ADP-ribose) Poly ADP ribose and the β domain of the DNA polymerase the tryptophan substitution in place of arginine, this change brings about the alteration in the binding efficiency either to both the domains or to one of domain [11]. In a study conducted by Przybylowska-Sygut K et al., and Moullan N et al., in polish population Arg194Trp was reported to be associated with breast cancer risk [11,26]. In a study conducted in white women found the Arg194Trp to be weakly associated with breast cancer risk, it was found to be associated with benign breast disease showing the heritability of the component [33]. Polymorphisms in XRCC1 have also been shown to lead to reduced capacity of DNA repair [34,35]. Fan X et al., Studies suggested that the Arg194Trp polymorphism in XRCC1 gene also has a key role in maintaining the efficiency of platinum-based chemotherapy [36,37]. Sanjari Moghaddam A et al., have propagated the idea that polymorphisms in XRCC1 are associated with a risk for breast cancer [38]. However, Al Mutairi FM et al., study on Saudi patients, observed that XRCC1 polymorphism, rs1799782, had a significant association with breast cancer risk [39]. A study from North eastern region of India reported significant association of Arg194Trp gene polymorphism with premenopausal breast cancer patients [40]. Th study conducted in southern India reported both the polymorphism to be associated with increased breast cancer risk [31]. Studies have also suggested the association of the polymorphism rs1799782 and rs25487 and the resistance capacity to cisplatin treatment in cancer patients [41,42]. The rs1799782 variant of XRCC1 did not show a significant association when comparing genotype distribution in healthy controls with breast cancer patients though the insilico analysis revealed deleterious effect of the protein and decreased activity, this may be attributed to other factors influencing the risk [Table/Fig-5].
A functional SNP rs25487 caused due to single base change from G to A results in substitution of glutamine aminoacid in place of arginine [9,10]. The A allele transition was found to be highly associated with increased levels of glycophorin mutation, elevated frequency of sister chromatid exchanges, higher DNA adduct formation and more sensitive towards ionising radiation exposures all of which are the responses towards decreased efficiency of base excision repair pathway. Several mutation in the XRCC1 gene have been associated with interruption in protein functionality, thereby the catalytic domain or the protein binding domain region is altered [43]. The functionally relevant polymorphism in XRCC1 gene rs25487 has been studied in various cancers. It has also been studied in breast cancer owing to its importance as it is located in COOH-terminal end of the PARP-binding BRCT-domain [7,44,45]. This single base change variation causes the substitution of alternate amino acid in the BRCT domain resulting in complete disturbance in the functioning of XRCC1 gene with low capability of DNA repair. Owing to its part in DNA repair machinery the variation due to SNP may modulate the susceptibility to breast cancer. Thus, when DNA repair proteins become deficient, probably due to genetic variants, this can lead to the initiation or can further aggravate breast cancer development. Despite the fact that many epidemiological studies have been performed in different populations on the association between DNA repair genes and breast cancer risk, only inconsistent and inconclusive results have been obtained. The XRCC1 (rs25487) did not show any significant association with breast cancer susceptibility in Jordanian population [46]. Recent meta-analysis conducted on the results obtained on 69 studies provided no evidence of association for breast cancer risk for the polymorphism rs25487 and rs179982 of the XRCC1 gene [47]. The results of the current study are also in agreement with those conducted by Wu K et al., a recent meta-analysis on 44 independent case-control studies [48]. The XRCC1rs25487 showed a significant association with hereditary and young breast cancer patients in Siberia [49]. The Non synonymous SNPrs25487 in the coding region of the XRCC1 gene revealed variations in the phenotype that affected the stability of the protein, function and structure that prohibit the protein from performing its function in terms of forming complex with DNA repair partners [Table/Fig-4,5]. Based on the insilico tools the extent of deletriousness of the variant protein is predicted. The study evaluated the risk attributed by the polymorphisms rs25487 and rs179982. The study suggests the involvement of rs25487 polymorphism as a susceptible marker for breast cancer in South Indian women. Genotype profiling enables individualised therapy further extensive study with large samples linking the DNA repair pathway, environment factors; ethnicity would shed more light on breast cancer susceptibility.
Limitation
The limitation of the present study was the its sample size consideration, a study with a larger sample size should be conducted in the future for the individual population.
Conclusion
The results obtained on the genotyping analysis for rs25487 was also similar in effect determined by stability prediction and pathogenicity analysis with computational tools revealing the nsSNP rs25487 interfering with function and structure of the protein. The rs1799782 variant of XRCC1 did not show a significant association with breast cancer risk. The XRCC1 gene has been implicated in breast cancer susceptibility in Southern Indian Women.
Five computational tools (I-Mutant Suite, iStable, PolyPhen2, SNAP, and PROVEAN) were used for insilico predictive analysis on the implication of the mutation on the function and structure of the variant XRCC1 protein
[1]. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Molecular portraits of human breast tumorsNature 2000 406:747-52.10.1038/3502109310963602 [Google Scholar] [CrossRef] [PubMed]
[2]. Malhotra GK, Zhao X, Band H, Band V, Histological, molecular and functional subtypes of breast cancersCancer Biol Ther 2010 10:955-60.10.4161/cbt.10.10.1387921057215 [Google Scholar] [CrossRef] [PubMed]
[3]. Mangia A, Malfettone A, Simone G, Darvishian F, Old and new concepts in histopathological characterization of familial breast cancerAnn Oncol 2011 22:24-30.10.1093/annonc/mdq66221285147 [Google Scholar] [CrossRef] [PubMed]
[4]. Liu ZQ, Zhang XS, Zhang SH, Breast tumor subgroups reveal diverse clinical prognostic powerSci Rep 2014 4:400210.1038/srep0400224499868 [Google Scholar] [CrossRef] [PubMed]
[5]. Higgins MJ, Baselga J, Targeted therapies for breast cancerJ Clin Invest 2011 121:3797-803.10.1172/JCI5715221965336 [Google Scholar] [CrossRef] [PubMed]
[6]. Toyokuni S, Akatsuka S, Pathological investigation of oxidative stress in the post-genomic eraPathol Int 2007 57:461-73.10.1111/j.1440-1827.2007.02127.x17610470 [Google Scholar] [CrossRef] [PubMed]
[7]. Hung RJ, Hall J, Brennan P, Boffetta P, Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE reviewAm J Epidemiol 2005 162:925-42.10.1093/aje/kwi31816221808 [Google Scholar] [CrossRef] [PubMed]
[8]. Pramanik S, Devi S, Chowdhary S, Surendran ST, Krishna Murthi K, Chakrabarti T, DNA repair gene polymorphisms at XRCC1, XRCC3, XPD, and OGG1 loci in Maharashtrian population of central IndiaChemosphere 2011 82:941-46.10.1016/j.chemosphere.2010.10.10021183201 [Google Scholar] [CrossRef] [PubMed]
[9]. Kohno T, Kunitoh H, Toyama K, Yamamoto S, Kuchiba A, Saito D, Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma riskCancer Sci 2006 97:724-28.10.1111/j.1349-7006.2006.00240.x16800823 [Google Scholar] [CrossRef] [PubMed]
[10]. Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Polymorphisms in the DNA repair gene XRCC1 and breast cancerCancer Epidemiol Biomarkers Prev 2001 10:217-22. [Google Scholar]
[11]. Przybylowska-Sygut K, Stanczyk M, Kusinska R, Kordek R, Majsterek I, Association of the Arg194Trp and the Arg399Gln polymorphisms of the XRCC1 gene with risk occurrence and the response to adjuvant therapy among Polish women with breast cancerClin Breast Cancer 2013 13:61-8.10.1016/j.clbc.2012.09.01923103366 [Google Scholar] [CrossRef] [PubMed]
[12]. Rowe BP, Glazer PM, Emergence of rationally designed therapeutic strategies for breast cancer targeting DNA repair mechanismsBreast Cancer Res 2010 12:20310.1186/bcr256620459590 [Google Scholar] [CrossRef] [PubMed]
[13]. Zhu Y, Hu J, Hu Y, Liu W, Targeting DNA repair pathways: a novel approach to reduce cancer therapeutic resistanceCancer Treat Rev 2009 35:590-96.10.1016/j.ctrv.2009.06.00519635647 [Google Scholar] [CrossRef] [PubMed]
[14]. Capriotti E, Fariselli P, Rossi I, Casadio R A, Three-state prediction of single point mutations on protein stability changesBMC Bioinformatics 2008 26(2):S610.1186/1471-2105-9-S2-S618387208 [Google Scholar] [CrossRef] [PubMed]
[15]. Chen CW LJ, Chu YW, iStable: off-the-shelf predictor integration forpredicting protein stability changesBMC Bioinformatics 2013 14(2):S510.1186/1471-2105-14-S4-S523514273 [Google Scholar] [CrossRef] [PubMed]
[16]. Calabrese R CE, Fariselli P, Martelli PL, Casadio R, Functionalannotations improve thepredictive score of human disease-related mutations inproteinsHum Mutat 2009 308:1237-44.10.1002/humu.2104719514061 [Google Scholar] [CrossRef] [PubMed]
[17]. Adzhubei IA SS, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, A method and server for predicting damaging missense mutationsNat Methods 2010 7:248-49.10.1038/nmeth0410-24820354512 [Google Scholar] [CrossRef] [PubMed]
[18]. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP, Predicting the functional effect of amino acid substitutions and indelsPLoS One 2012 7(10)10.1371/journal.pone.004668823056405 [Google Scholar] [CrossRef] [PubMed]
[19]. Bjelland S, Seeberg E, Mutagenicity, toxicity and repair of DNA base damage induced by oxidationMutat Res 2003 531:37-80.10.1016/j.mrfmmm.2003.07.00214637246 [Google Scholar] [CrossRef] [PubMed]
[20]. Chao MW, Kim MY, Ye W, Ge J, Trudel LJ, Belanger CL, Genotoxicity of 2,6- and 3,5-dimethylaniline in cultured mammalian cells: the role of reactive oxygen speciesToxicol Sci 2012 130(1):48-59. [Google Scholar]
[21]. Thompson LH, Schild D, Homologous recombinational repair of DNA ensures mammalian chromosome stabilityMutat Res 2001 477:131-53.10.1016/S0027-5107(01)00115-4 [Google Scholar] [CrossRef]
[22]. Thompson PA, DeMarini DM, Kadlubar FF, McClure GY, Evidence for the presence of mutagenic arylamines in human breast milk and DNA adducts in exfoliated breast ductal epithelial cellsEnviron Mol Mutagen 2002 39:134-42.10.1002/em.1006711921181 [Google Scholar] [CrossRef] [PubMed]
[23]. Santella RM, Gammon MD, Zhang YJ, Motykiewicz G, Young TL, Hayes SC, Immunohistochemical analysis of polycyclic aromatic hydrocarbon-DNA adducts in breast tumor tissueCancer Lett 2000 154:143-49.10.1016/S0304-3835(00)00367-0 [Google Scholar] [CrossRef]
[24]. Gammon MD, Sagiv SK, Eng SM, Shantakumar S, Gaudet MM, Teitelbaum SL, Polycyclic aromatic hydrocarbon-DNA adducts and breast cancer: a pooled analysisArch Environ Health 2004 59:640-49.10.1080/0003989040960294816789472 [Google Scholar] [CrossRef] [PubMed]
[25]. Rundle A, Tang D, Zhou J, Cho S, Perera F, The association between glutathione S-transferase M1 genotype and polycyclic aromatic hydrocarbon-DNA adducts in breast tissueCancer Epidemiol Biomarkers Prev 2000 9:1079-85. [Google Scholar]
[26]. Moullan N, Cox DG, Angèle S, Romestaing P, Gérard JP, Hall J, Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapyCancer Epidemiol Biomarkers Prev 2003 12:1168-74. [Google Scholar]
[27]. Ratnasinghe D, Yao S-X, Tangrea JA, Qiao Y-L, Mark R, Polymorphisms of the DNA repair gene XRCC1 and lung cancer riskCancer Epidem Biomar 2001 10:119-23. [Google Scholar]
[28]. Sturgis EM, Spitz MR, Wei Q, Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neckCarcinogenesis 1999 20:2125-29.10.1093/carcin/20.11.212510545415 [Google Scholar] [CrossRef] [PubMed]
[29]. Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, Case LD, Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivityCarcinogenesis 2001 22:917-22.10.1093/carcin/22.6.91711375899 [Google Scholar] [CrossRef] [PubMed]
[30]. Försti A, Angelini S, Festa F, Sanyal S, Zhang Z, Grzybowska E, Pamula J, Single nucleotide polymorphisms in breast cancerOncol Rep 2004 11:917-22.10.3892/or.11.4.91715010895 [Google Scholar] [CrossRef] [PubMed]
[31]. Chacko P, Rajan B, Joseph T, Mathew BS, Pillai MR, Polymorphisms in DNA repair gene XRCC1 and increased genetic susceptibility to breast cancerBreast Cancer Res Treat 2005 89:15-21.10.1007/s10549-004-1004-x15666192 [Google Scholar] [CrossRef] [PubMed]
[32]. Saadat M, Kohan L, Omidvari S, Genetic polymorphisms of XRCC1 (codon 399) and susceptibility to breast cancer in Iranian women, a case-control studyBreast Cancer Res Treat 2008 111:549-55.10.1007/s10549-007-9811-517987379 [Google Scholar] [CrossRef] [PubMed]
[33]. Smith TR, Miller MS, Lohman K, Lange EM, Case LD, Mohrenweiser HW, Polymorphisms of XRCC1 and XRCC3 genes and susceptibility to breast cancerCancer Letter 2003 190:183-90.10.1016/S0304-3835(02)00595-5 [Google Scholar] [CrossRef]
[34]. Vodicka P, Stetina R, Polakova V, Tulupova E, Naccarati A, Vodickova L, Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjectsCarcinogenesis 2007 28:657-64.10.1093/carcin/bgl18717028303 [Google Scholar] [CrossRef] [PubMed]
[35]. Au WW, Salama SA, Sierra-Torres CH, Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assaysEnviron Health Perspect 2003 111:1843-50.10.1289/ehp.663214630517 [Google Scholar] [CrossRef] [PubMed]
[36]. Fan J, Wilson DM, Protein-protein interactions and posttranslational modifications in mammalian base excision repairFree Radic Biol Med 2005 38:1121-38.10.1016/j.freeradbiomed.2005.01.01215808410 [Google Scholar] [CrossRef] [PubMed]
[37]. Fan X, Xiu Q, Effect of X-ray repair cross complementing group 1 polymorphisms on the efficacy of platinum-based chemotherapy in patients with nonsmall cell lung cancerJ Cancer Res Ther 2015 11:571-74.10.4103/0973-1482.15908526458583 [Google Scholar] [CrossRef] [PubMed]
[38]. Sanjari Moghaddam A, Nazarzadeh M, Sanjari Moghaddam H, Bidel Z, Keramatinia A, Darvish H, XRCC1 gene polymorphisms and breast cancer risk: a systematic review and meta- analysis studyAsian Pac J Cancer Prev 2016 17:323-30.10.7314/APJCP.2016.17.S3.32327165246 [Google Scholar] [CrossRef] [PubMed]
[39]. Al Mutairi FM, Alanazi M, Shalaby M, Alabdulkarim HA, Pathan AA, Parine NR, Association of XRCC1 gene polymorphisms with breast cancer susceptibility in Saudi patientsAsian Pac J Cancer Prev 2013 14:3809-13.10.7314/APJCP.2013.14.6.380923886187 [Google Scholar] [CrossRef] [PubMed]
[40]. Rekha K, Devi JA, Kanwar Narain, DNA Repair Mechanism Gene, XRCC1A (Arg194Trp) but not XRCC3 (Thr241Met) Polymorphism Increased the Risk ofBreast Cancer in Premenopausal Females:A Case-Control Study in NortheasternRegion of IndiaTechnology in Cancer Research & Treatment 2017 16(6):1150-59.10.1177/153303461773616229332455 [Google Scholar] [CrossRef] [PubMed]
[41]. Jin ZY, Zhao XT, Zhang LN, Wang Y, Yue WT, Xu ST, Effects of polymorphisms in the XRCC1, XRCC3 and XPG genes on clinical outcomes ofplatinum-based chemotherapy for treatmentof non-small cell lung cancerGenet Mol Res 2014 :7617-25.10.4238/2014.March.31.1324737519 [Google Scholar] [CrossRef] [PubMed]
[42]. Kudo K, Gavin E, Das S, Amable L, Shevde LA, Reed E, Inhibition of Gli1 results in altered cJunactivation, inhibition of cisplatin-inducedupregulation of ERCC1, XPD and XRCC1, andinhibition of platinum-DNA adduct repairOncogen 2012 31:4718-24.10.1038/onc.2011.61022266871 [Google Scholar] [CrossRef] [PubMed]
[43]. Caldecott KW, Aoufouchi S, Johnson P, Shall S, XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly ADP-ribose polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitroNucleic Acids Res 1996 24:4387-94.10.1093/nar/24.22.43878948628 [Google Scholar] [CrossRef] [PubMed]
[44]. Shen MR, Jones IM, Mohrenweiser H, Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humansCancer Research 1998 58:604-08. [Google Scholar]
[45]. Masson M, Niedergang C, Schreiber VER, Muller S, Menissier-de Murcia J, XRCC1 is specifically associated with poly (ADP-ribose) polymerase and negatively regulates its activity following DNA damageMolecular and Cellular Biology 1998 18:3563-71.10.1128/MCB.18.6.35639584196 [Google Scholar] [CrossRef] [PubMed]
[46]. Zoubi MSA, X-ray repair cross-complementing protein 1 and 3 polymorphisms and susceptibility of breast cancer in a Jordanian populationSaudi Med J 2015 36(10):1163-67.10.15537/smj.2015.10.1265926446325 [Google Scholar] [CrossRef] [PubMed]
[47]. Qiao L, Feng X, Wang G, Zhou B, Yantong Y, Li M, Polymorphisms in BER genes and risk of breast cancer: evidences from 69 studies with 33760 cases and 33252 controlsClinical Research Paper 2018 9(22):16220-33.10.18632/oncotarget.23804 [Google Scholar] [CrossRef]
[48]. Wu K, Su D, Lin K, Luo J, Au WW, XRCC1 Arg399Gln gene polymorphism and breast cancer risk: a meta-analysis based on case-control studiesAsian Pac J Cancer Prev 2011 12:2237-43. [Google Scholar]
[49]. Krivokuca AM, Cavic MR, Malisic EJ, Rakobradovic JD, Kolarevic-Ivankovic D, Tomasevic ZI, Polymorphisms in cancer susceptibility genes XRCC1, RAD51 and TP53 and the risk of breast cancer in Serbian womenInt J Biol Markers 2016 31(3):e258-63.10.5301/jbm.500020126954070 [Google Scholar] [CrossRef] [PubMed]