Hypothyroidism is a common endocrinological disorder in India, it controls the metabolism and developmental processes in human [1,2]. Various other metabolic disorders like obesity and dyslipidemia can complicate hypothyroidism [3]. Genetic predispositions of these complications of hypothyroidism are having an obvious effect through out the life of a patient [4]. Some observational studies have suggested that hypothyroid patients have accelerated coronary atherosclerosis [5]. Hypothyroidism is one of the most common associated factor for the development of secondary hypercholesterolaemia [6]. More than 90% of hypothyroidism patients are hyperlipidemic [7]. Al-Tonsi AA et al., has shown a significant rise in fasting Total Cholesterol (TC), Triglyceride (TG) and LDLC concentrations with declining thyroid functions [8]. Furthermore, overt hypothyroidism also affects HDLc levels [9]. Bhalodkar NC et al., observed higher TG/HDL ratio (>3.8) in Asian Indians which may predispose high prevalence rate of atherogenesis [10]. Same results were observed by some authors. [11]. Association studies have identified several genetic variants related to Thyroid Hormone Receptor α (THRα) and Thyroid hormone receptor β (THRβ) which are located on chromosome no 17 and chromosome no 3 respectively [12]. Resistance in THRα gene causes hypothyroidism along with the significant alteration in T3/T4 ratio [13]. SNP of any of these genes in hypothyroid patients can alter the quality of life. Furthermore they can make significant consequences like obesity, dyslipidemia, coronary atherosclerosis and defective cognition [4]. It is being observed that mutation in THRα gene leads to erythroid disorders in abnormal red blood cell indices [14].
Few authors documented the link between THRα polymorphism rs-939348 and higher Systolic Blood Pressure (SBP) with more risk of hypertension [15]. Significant correlation between THRα polymorphism rs-939348 and central obesity was also noticed by some authors [16]. Same restriction site polymorphism was also detected in thyroid cancer [17].
Implication of THRα polymorphism rs-939348 on dyslipidemia is unclear. Given the limited information concerning the implication of THRα gene involved with lipid profile, the study analysed the risk association between genetic variant of THRα rs-939348 and atherogenic dyslipidemia.
Materials and Methods
This cross-sectional, observational, hospital based study was conducted in the Department of Biochemistry, Calcutta National Medical College and Hospital, Kolkata, in the period between 15.01.17 to 15.09.17. A total of 191 patients of Primary hypothyroidism (M=91, F=100) ranging from 25 to 65 years of age were selected from Thyroid clinic of Biochemistry Department. All participants were included with fT4 < 0.8 ngm/dL and TSH > 4.7 μIU/mL. All participants were recruited with their written consents.
Excluded from this study were those with diabetes mellitus, hypertension, active neoplasm or history of neoplasm, severe liver dysfunction, severe renal failure (Stage III & IV), nephritic/nephrotic syndrome, diseases of the pituitary gland or hypothalamus including secondary hypothyroidism, major surgery within two weeks of enrolment, a severe psychiatric condition not related to hypothyroidism symptoms, GIT malabsorption diseases, pregnancy, alcohol abuse, concurrent medication that may interfere with thyroid hormone absorption or activation, patients taking oral contraceptive pills, hormone replacement therapy, and oral steroids, critically ill patients, Hashimoto’s thyroid disease and thyroidectomized patients on L-T4 treatment. Patients with history of smoking were also excluded from the study.
The whole process strictly followed the guidelines and regulations set by Helsinki Declaration of 1975, with all amendments and revisions. The study was approved by Institutional Ethics Committee Application no – CNMC/10, dated on 15.03.16.
Sample Collection and Handling
Fasting blood samples were collected from patients who matched the study criteria. Blood (5 mL) was withdrawn and distributed into anticoagulant – free plain tube (2 mL) and EDTA tube (3 mL). The blood sample in the plain tube was centrifuged after 30 minutes of sampling and serum was isolated and stored at -20°C and sent to the laboratory for biochemical analysis. Assays include TSH, fT4, (S) Triglyceride and (S) HDLc. Biochemical parameters of plain tube were estimated on the same day. EDTA tubes were stored properly at -20o C.
fT4 estimation were done by competitive ELISA [18] (Aspen Laboratory Pvt., Ltd.,). TSH was analysed by Sandwitch ELISA [19] (Aspen Laboratory Pvt., Ltd.,) in a standard ELISA reader (ERBA, Lisascan EM) (S) Triglyceride and HDLc were estimated by GPO/PAP [20] and PEG/CHOD-PAP [21] methods respectively. Assays were carried out using the Konelab Automated analyser Prime 601i (Thermo Fisher Scientific, USA).
Samples were assayed in duplicate and the mean of the paired results was determined. Patients diagnosed as hypothyroids were divided into two groups. One group with high triglyceride and low HDLc levels, another group with normal range of these two parameters.
Blood collected in the EDTA tubes was used for DNA extraction.
Molecular Analysis
DNA extraction: The venous blood, which was collected in the evacuated EDTA tubes, was used for DNA extraction. DNA from study subjects was isolated from peripheral blood (EDTA sample) using the standard phenol–chloroform method [22]. The integrity of genomic DNA was tested by resolving DNA extracts on a 0.8% agarose gel by electrophoresis (Low EEO, SRL), followed by visualisation with ethidium bromide staining. A quantitative spectrophotometric assay of DNA was performed with a MS-UV+ spectrophotometer (Motras Scientific, New Delhi, India). Wavelengths of 260 and 280nm were used to measure the absorbance (A260 and A280, respectively). The absorbance quotient (OD260/OD280) signifies the purity of DNA. An OD value in between 1.8 < ratio (R) <2.0 indicates the purified DNA for PCR. The good samples were then transferred to a fresh tube, and 1 μl of 10 μg/ml RNase A (Chromus Biotech) was added. The sample was incubated at 37°C for one hour to improve the purity. The samples were stored at −20 °C until use.
PCR-RFLP
PCR/RFLP: SNPs, PCR primer sequence and restriction enzymes were resourced from UCSC Genome Bioinformatics Site and Primer3, NEB cutter programs, respectively [23-26].
The genomic DNA was amplified using the following steps: Denaturation of double stranded genomic DNA at 94°C for 5 minutes, DNA amplification using 30 cycles. Each cycle consists of: denaturation at 94°C for 30 seconds, annealing at 54°C for 30 seconds, extension at 72°C for 40 seconds, final elongation at 72°C for 7 minutes, and ending reaction at 4°C. For SNP rs939348, e PCR primers, F:5’ CCTGTGTCTCCCAGCTTAGG 3’R: 5’CCACCAGACTCACAGCCTCT 3’, the PCR product is 190 bp(Size of amplified fragment), the sizes of the digested fragments of the examined SNPs are C allel 190 T allel 48 and 142 bp. The restriction enzyme (Mse I) digestion was carried out in 1 μl containing 10 units of enzyme (Mse I) with 5 μl of 10X buffer and 1 μg of PCR product and incubated at 37°C for 30 minutes. Restriction enzyme was obtained from New England (BioLabs). Size of restricted fragments for C allele is 190 bp and T allele is 142 and 48 bp. PCR products and digested fragments were detected using electrophoresis on 1.2% and 2% agarose respectively.
Statistical Analysis
Data was analysed using the SPSS 17 software. Continuous variables were expressed as mean (standard deviation) and the differences were accomplished by comparison via student’s unpaired 2-sided t-test or one way ANOVA as appropriate. Discrete variables were expressed as counts and frequencies and were compared using the chi-square test. If N<5, exact fisher statistics was used, the genotype and allele frequencies of examined SNPs (for rs 939348) were determined. The genotype distributions of SNPs were analysed in agreement with the Hardy-Weinberg equilibrium. A significant difference was considered at p <0.05.
Results
[Table/Fig-1] shows mean age, TSH, fT4, triglyceride and HDLc levels in the groups. Only triglyceride and HDLc concentrations were significant (p<0.0001) between the groups.
Demographic, biochemical and hormonal characteristics of the studied group.
| NormolipidemicHypothyroid (n=99) mean±SD | DyslipidemicHypothyroid (n=92) mean±SD | p-value |
|---|
| Age (years) | 45.3±8.1 | 45.10±7.9 | <0.8632 |
| Gender (M/F) | 46/53 | 45/47 | |
| TSH (uIU/ml) | 14.81±6.78 | 14.88±7.40 | <0.9457 |
| fT4 (ng/dl) | 0.71±.07 | 0.7±0.23 | <0.68 |
| Triglyceride (mg/dl) | 125.35±36.84 | 331.13±122.58 | <0.0001 |
| HDL (mg/dl) | 45.11±8.15 | 35.73±4.72 | <0.0001 |
[Table/Fig-2] shows genotypes and allele frequencies in two groups. As regards rs -939348 polymorphism, the genotype frequencies in hypothyroid patients without dyslipidemia were as follows: CC genotype was 72/99 (72.72%), CT genotype was 16/99(16.16%) and TT genotype was 4/99(4.04%), whereas hypothyroid patients having dyslipidemia the CC, CT and TT genotype frequencies are 48/92(52.17%),35/92(38.04%) and 16/92(17.39%) respectively.
THR α Polymorphism genotype and allele frequencies in the groups.
| Genotypes | NormolipidemicHypothyroid n(%)(N=99) | DyslipidemicHypothyroid n(%) (N=92) | Odds Ratio | 95% CI | p |
|---|
| SNP THR α rs939348 | C/C | 72(72.72%) | 48(52.17%) | 2.44 | 1.33- 4.46 | 0.004 |
| C/T | 16(16.16%) | 35(38.04%) | 0.31 | 0.15- 0.62 | 0.0009 |
| T/T | 4(4.04%) | 16(17.39%) | 0.20 | 0.06- 0.62 | 0.0036 |
| AlleleC Allele | 160(86.96%) | 131(66.16%) | 3.40 | 2.02- 5.7 | <0.0001 |
| T Allele | 24(13.04%) | 67(33.84%) | 0.2933 | 0.17- 0.49 | <0.0001 |
The study found there was a definite declining trend of CC genotypes in dyslipidemic hypothyroids when compared to their normolipidemic partners.(Odds Ratio 2.44, 95% CI 1.33- 4.46, p=0.004) whereas the opposite trend were found in case of CT (Odds ratio 0.31, 95% CI 0.15 -0.62, p=0.0009) and TT (Odds ratio 0.2, 95%CI 0.06-0.62 p=0.0036), as regards allele frequency, C allele was significantly decreased (Odds ratio 3.4, 95% CI 2.02-5.7, p<0.0001) whereas T allele was significantly increased (Odds ratio 0.29, 95% CI 0.17-0.49, p<0.0001) in dyslipidemic hypothyroids than normolipidaemics.
Furthermore, the study found a trend of increasing T allele in dyslipidemic hypothyroids. This indicates significant association of mutant T allele with developing dyslipidemia in hypothyroids and C allele may be considered as protective.
[Table/Fig-3] shows that in dyslipidemic hypothyroids, triglyceride level was different in different genotype (p=0.02), and allele (p=0.04) groups whereas HDLc is different in different genotype (p<0.0001) but not in different allele (p=0.28) groups.
Biochemical and hormonal parameters in different THRα rs939348 polymorphisms genotypes and alleles in the groups.
| THR α genotypes | | THR α alleles | |
|---|
| C/C | C/T | T/T | p | C allele | T allele | p |
|---|
| Normolipidemic Hypothyroid |
| TG(mg/dl) | 123.81±36.38 | 121.97±35.86 | 133.56±40.40 | 0.566 | 123.32±35.97 | 125.60±37.66 | 0.67 |
| HDL(mg/dl) | 44.50±1.88 | 45.4±1.91 | 44.47±1.73 | 0.071 | 44.74±1.4 | 45.10±1.0 | 0.06 |
| Dyslipidemic Hypothyroid |
| TG(mg/dl) | 280.62±84.59 | 388±88.56 | 339.19±89.38 | 0.02 | 316.41.±85.91 | 355.46±89.10 | 0.04 |
| HDL(mg/dl) | 35.70±0.6 | 33.35±0.89 | 36.0±1.09 | <0.0001 | 34.91±0.69 | 35.11±1.56 | 0.28 |
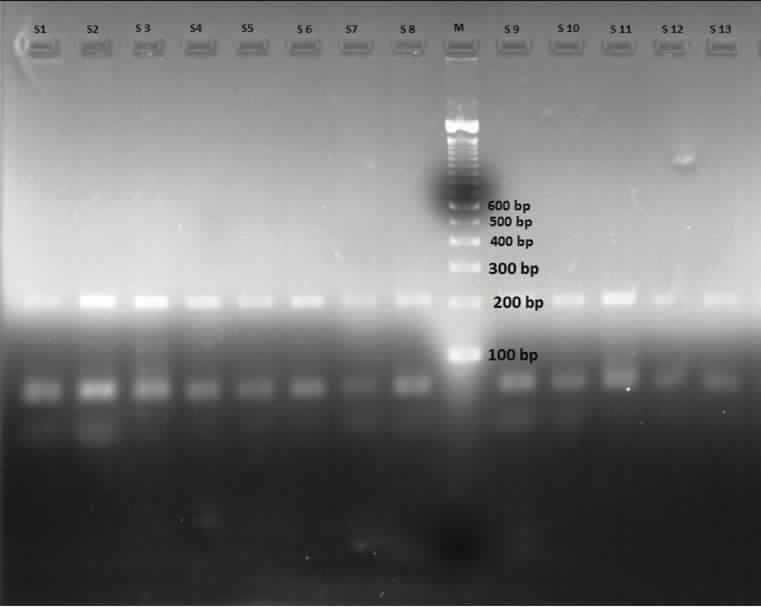
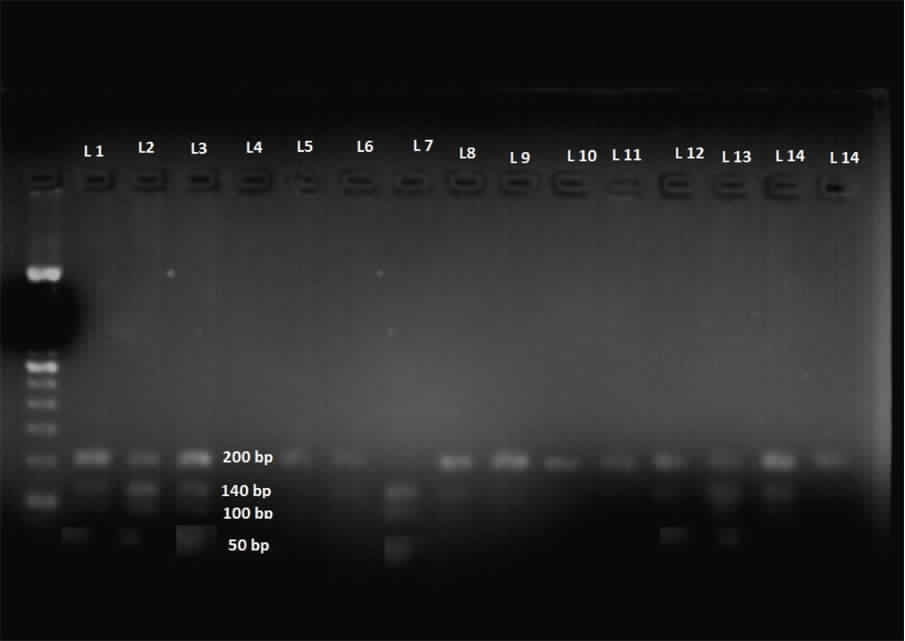
[Table/Fig-4] shows the agarose gel electrophoresis showing PCR products and [Table/Fig-5] shows the agarose gel electrophoresis after restriction and digestion of PCR products.
Agarose gel electrophoresis showing PCR products.
PCR products (size 190 bp) of rs 939348 of THR α gene – S- sample, M- ladder.
Agarose gel electrophoresis after restriction and digestion of PCR products.
Restriction fragment products of those samples are being marked as (L); size of C allele – 190 bp and T allele-142 & 48 bp. Lane 1,2,3 represent heterozygote alleles, Lane 7 represents homozygote allele. Ladder is in extreme left
Discussion
Since there is a lack of genome wide association study, between THRα SNP and altered lipid profile, a population gene approach like present study can help to detect associations between SNP and dyslipidemia in hypothyroids.
The study tried to analyse the association between SNP rs 939348 of THRα gene with altered lipid profile among hypothyroid patients. The study observed higher fasting TG and lower HDLc levels among T/T and C/T genotypes in comparison to C/C.
Goumidi L et al., demonstrated the association between THR α rs-939348 polymorphism and SBP, furtherrmore, they have also shown that THR α rs 939348 SNP was associated with 25% higher risk of hypertension [15]. Al-Azzam SI et al., also observed the link between the same THRα restriction site and increased waist circumference in hypothyroids. They concluded with the fact that hypothyroid patients with T/T genotype of rs-939348 of THR α gene had more waist circumference than other genotype [16]. These two studies indicated that T/T genotypes and T alleles are more vulnerable to get the diseases. The former also found a significant higher risk of developing Alzeimers Disease in T/T subjects of rs-939348 SNP of THRα, when compared with C allele bearers [27]. Pastor S et al., observed THRα rs939348 was found to be associated with higher risk of differential thyroid cancer [17]. Another two cohort studies on THRα, one had SNP rs-12939700, showed high risk of CVD (cardio vascular disease). The 2nd one in a normal Spanish cohort, observed SNP at rs-15684000. They had higher TC, fasting TG, BMI and larger waist circumference. In a subgroup of same normal subjects, six years later, increased risk of developing obesity was found [28]. In correspondence with these studies, the study found higher TGL and lower HDLc levels among T/T and C/T genotype in comparison to C/C.
In hypothyroidism, the low activity of Cholesterol Ester Transfer Protein (CETP) and hepatic lipase causes diminished transport of cholesteryl ester from HDL2 to VLDL, IDL and HDL3. Diminished LPL activity results in diminished conversion of VLDL into LDL and predisposes the increase of the TG, IDL, VLDL and small dense highly atherogenic particles [29].
Thyroid hormone regulates TG homeostasis which is influenced by APOA5 gene transcription. APOA5 activates Lipoprotein Lipase (LPL) mediated TG metabolism and inhibiting hepatic VLDL to TG formation.
Patients with SNP or mutation of APOA5 manifest markedly reduced plasma post heparin LPL activity and hypertriglyceridemia [30].
THRα has been related to adipocyte growth, documented by some authors [31]. THRα isoform is highly expressed in adipocytes, implicates that THRα seems to be involved in lipid accumulation in adipocytes [32]. Gene specificity was not obvious, rather specific intracellular modification of receptors was indicated more significant.
Authors arouse attention to the splicing of THRα transcripts, partially depends on location of SNP at restriction site. Altered splicing site possibly influences in THRα transcripts [31].
SNP rs-939348 is located in the 1st intron after translation start site; it could modulate the level of expression of one of the transcripts encoded by THRα gene [15]. Al-Azzam SI et al., suggested that this SNP could have a role in the development of metabolic syndrome and Coronary Artery Disease (CAD). They indicated TT genotype and T allele are mainly responsible [16]. T allele of rs-939348 SNP creates a potential binding site for glucocortocoid receptors or Hepatic Nuclear Factor 1 (HNF 1). These transcription factors could regulate THRα gene expression. Otherwise these polymorphisms could influence the mRNA stability or splicing balance between the active THRα 1isoform and inactive other isoform [27].
Above observations supported our study in the way that high TG and low HDL C levels were found among C/T and T/T genotype frequency of SNP rs-939348 of THRα gene. The present study observed the trend of increasing T allele in dyslipidemic hypothyroids. This indicates significant association of mutant T allele with developing dyslipidemia in hypothyroids and C allele may be considered as protective.
Limitation
Small sample size and duration may interfere the results of the study. Furthermore, the study selected the scattered population attending a tertiary care hospital. Larger sample size may be beneficial.
Conclusion
The present study constituted as an association study of THRα gene polymorphism and atherogenic dyslipidemia risk. The study also observed association between C/T or T/T genotype of THRα gene rs939348 and higher TG with lower HDLc levels. It can be hypothesised from the study that C/C genotype may have a lower incidence of dyslipidemia.