Microorganisms which are distinguished by antigenic differences are known as serotypes. The different serotypes are identified using serotyping [2]. Characterization of serotypes can be done by conventional and molecular methods. In general, conventional method has lower discriminatory power than molecular methods. Most molecular methods require costly equipment, reagents but are fast to perform and have capability of differentiating wide variety of strains [3]. Co-A, a conventional method used to characterize K antigen, differentiates isolates into many serotypes. Antiserum required in the method was raised in rabbit. Though the raising and maintenance of the antiserum, the Co-A method is difficult to handle, laborious and time consuming [4], it is cheap, sensitive, and quick in diagnosis, processed without any requirement of expensive equipments and experts. Antiserum once prepared is very long lasting so this method was opted.
The spread of drug resistant strains is plasmid mediated and these may also contain virulence determinants to make them an effective pathogen [8]. So, the present study was planned to know the occurrence of hvKP, to understand the incidence of K1, K2, K3 and K4 serotypes of K. pneumoniae and to study the occurrence of beta- lactamase genes among the K1, K2, K3 and K4 positive serotypes of K. pneumoniae.
Materials and Methods
Sample Collection: A prospective study was carried out at the Department of Microbiology, KVG Medical College and Hospital, Sullia, Karnataka, India. Various consecutive, non-duplicate isolates of K. pneumoniae recovered from clinical specimens such as Urine (n=38), sputum (n=18), pus (n=1), blood (n=1) and others (n=2) obtained from both outpatients and inpatients attending KVG Medical College, Sullia between April to December 2017 were included in the study. The minimum sample size was calculated using the following formula.
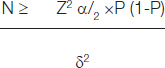
Level of significance is 1.96, δ2 = (0.05)2, The estimated prevalence is 6% (ESBL-producers among the serotype K1). So P =0.06
Hence, the minimum sample size was 60
Klebsiella pneumoniae isolated without any clinical disease, suggesting colonization were excluded from the study. Approval from the KVG Medical College Institutional Scientific and Ethics Committee was obtained (IEC/ 01/2016).
Phenotypic Identification: Biochemical characterization of the isolates of Klebsiella collected from various clinical specimens was done by applying standard identification procedures [9].
Co-Agglutination Test [10]:
Preparation of Purified Capsular Polysaccharide (CPS): The reference capsular serotypes of K. pneumoniae procured from MTCC Chandigarh were grown on Luria broth and incubated at 37°C. Further growth of organisms in Luria broth was stopped by adding cetavlon to a final concentration of 0.5% (wt/vol); kept at room temperature by constant stirring for 30 minutes and then centrifuged at 10000 X g for 15 minutes to get precipitate. Equal amount of 1M CaCl2 solution and 25% ethanol was added to the precipitate and stirred well for 60 minutes at room temperature. The precipitated nucleic acid was removed by centrifuging at 10,000 X g for 30 minutes. To the supernatant 80% ethanol was added to get the precipitated CPS. Then the CPS was extracted by treating with an equal volume of chloroform butanol solution in a proportion of 5:1. The water phases were dialyzed against distilled water at 40°C. Lipopolysaccharide was removed by centrifuging the solution at 100,000 X g for 16 hours. The supernatant fluid which contains CPS was collected and analysed for its purity by colorimetric assay.
Antisera Preparation: Rabbits were immunized to get hyper immune sera against purified CPS antigen. Two milliliter (mL) of complete Freund’s adjuvant was added to 2 ml of 0.25 mg/ml purified CPS in Phosphate Buffered Saline (PBS). Mixed well and stored at 4°C. About 0.1 ml of CPS, adjuvant mixture was administered subcutaneously. Subsequent Booster immunizations were done by injecting Incomplete Freund’s adjuvant /antigen emulsion following the same procedure as above. The first booster immunization was administered 4 weeks after the priming immunization. After 10 days blood sample was collected from animal and serum was prepared [11].
(The Ref. capsular strains – Klebsiella pneumoniae subsp.ozaenae (ATCC® 11296™) Klebsiella pneumoniae subsp. rhinoscleromatis (ATCC® 13884™) Staphylococcus aureus subsp. aureus (ATCC® 12598™) was procured from MTCC Chandigarh).
Preparation of S. aureus Cowan 1 Strain Reagent: To find out K antigens of K.pneumoniae by Co-A method using S. aureus Cowan 1 strain, the following procedure was used. Culture of S. aureus Cowan 1 strain was washed with PBS, kept at room temperature for three hours, washed with PBS and adjusted the concentration up to 10% with PBS. It was again heated at 80°C for 15 minutes in water bath and washed and adjusted 10% concentration with PBS and stored at 4°C. To 0.025 ml of antiserum 1ml of stored suspension of S. aureus Cowan 1 strain was added and mixed well, allowed to stand for some time and then washed twice with phosphate buffered saline and stored and used for at least for one year of duration [12].
Preparation of Antigen: Overnight cultures of K. pneumoniae (test strains) were added with 2ml saline, mixed and kept at room temperature for one hour. Centrifuged – clear supernatant was used as capsular antigen.
Test Procedure: One drop of S. aureus reagent was mixed with equal volume of antigen. The presence of agglutination suggested capsular antigen [12].
Detection of hvKP: String test was used to detect hvKP strains of K. pneumoniae as per the procedure followed by Fang CT., et al [13]. The appearance of viscous strings of >5 mm in length while a loop was stretched on the colonies of K. pneumoniae, suggested positive string test.
Detection of ESBL Production:
Screening of ESBL production: Disk diffusion test was used to know the resistance to third generation cephalosporins such as cefotaxime, ceftazidime, ceftriaxone as per the CLSI guidelines 2017 [14].
Confirmation of ESBL detection by Double Disc Synergy Test (DDST): An augmentation of zones of Disks containing cephalosporins 30μg (cefotaxime, ceftazidime) by clavulanic acid (10 μg) kept at a distance of 20mm centre-to-centre between cephalosporins and clavulanic acid suggested the possible presence of an ESBL producing serotype [15].
Escherichia coli (ATCC® 25922™) was used as ESBL negative control and Klebsiella pneumoniae subsp. pneumoniae (ATCC® 700603™) as positive control.
Detection of K1 and K2 serotypes and ESBL genes by Polymerase Chain Reaction (PCR): Extraction of DNA among K.pneumoniae serotypes were done as per the procedure given by Bora A et al., [16].
Nuclease-free water was included as negative control Klebsiella pneumoniae subsp. pneumoniae ATCC® 700603™ was used as a positive control for blaSHV. A previously confirmed K. pneumoniae isolate possessing blaTEM and blaCTX-M genes were used as positive control for blaTEM and blaCTX-M genes. The total concentration of PCR reaction to do single test was 25 μL {PCR ready mix -12.5 μL (Sigma Aldrich, Merck’s Life Science), Forward primer-1μl, Reverse primer-1 μL (Bioserve Hyderabad, India)}.
K1 F (5′-GTAGGTATTGCAAGCCATGC -3′) R (5′-GTAGGTATTGCAAGCCATGC-3′) 1283bp [17] K2 F (5′ – GGAGCCATTTGAATTCGGTG-3′)
R (5′- TCCCTAGCACTGGCTTAAGT-3′) 646bp [17] TEM F (5′ – AAAATTCTTGAAGACG -3′) R (5′- TTACCAATGCTTAATCA -3′) 1080bp [18]
SHV F (5′ – ATTTGTCGCTTCTTTACTCGC-3′) R (5′- TTATGGCGTTACCT TTG ACC-3′) 1018bp [19] CTX-M1 F (5′ – AAAAATCACTGCGCCAGTTC -3′)
R (5′- AGCTTATTCATCGCCACGTT-3′) 445 bp [20], Template DNA- 4 μL and Nuclease free water - 6.5 μL (Hi Media Laboratories Pvt., Ltd., Mumbai, India). Other consumables used: 100 bp DNA ladder (Hi Media Laboratories,), 50x TAE buffer (Hi Media), 6x loading dye solution (Hi Media), Ethidium bromide solution (Sigma Aldrich).
Detection of ESBL genes such as blaTEM,blaSHV and blaCTX-M was done among the capsular serotypes using above mentioned primers and standardized PCR cycling conditions and procedures [Table/Fig-1].
Standardized PCR cycling conditions.
| Beta lactamase | Initial denaturation | Denaturation | Annealing | Elongation | Final elongation |
|---|
| K1 antigen | 950C for 4 min | 940Cfor 30 sec30 cycles | 570C for 30 sec30 cycles | 720C for 30 sec30 cycles | 720C for five min |
| K2 antigen | 940C for 3 min | 940C for 2 min28 cycles | 650C for 1min28 cycles | 720C for 1min28 cycles | 720C for seven min |
| bla TEM | 950C for 2 min | 950C for 45 sec30 cycles | 620C for 45 sec30 cycles | 720C for 60 sec30 cycles | 720C for five min |
| bla SHV | 940C for 2 min | 940C for 40 sec35 cycles | 580C for 40 sec35 cycles | 720C for 60 seconds35 cycles | 720C for seven min |
| CTX-M-1 | 940C for 1 min | 940C for 40 sec35 cycles | 550C for 40 sec35 cycles | 720C for 60 seconds35 cycles | 720C for seven min |
All the three genes were detected by uniplex PCR using Thermocycler (Quanta Biotech, 96 S). PCR run conditions were programmed using the supplied software.
Statistical Analysis
Chi-square test was used to compare Co-A and PCR study. Level of significance for this study was 5%. Software used was SPSS version 16.0.
Results
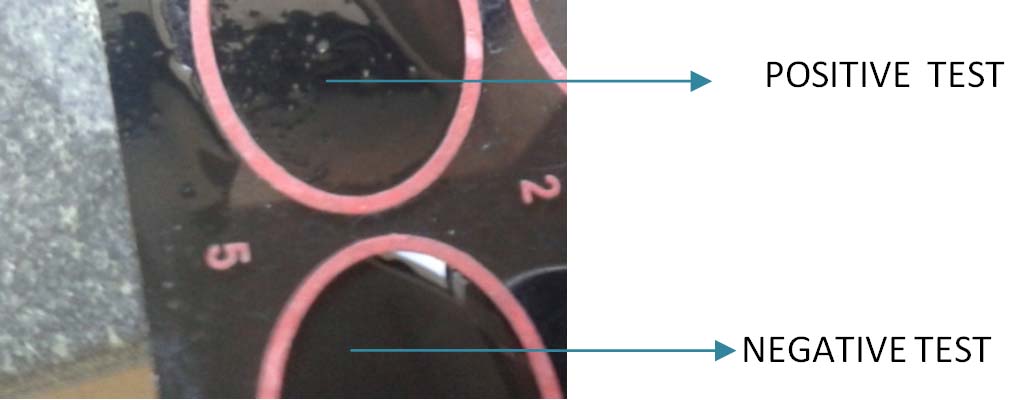
Rate of typability by Co-A was found to be 13(21.67%), 0 (0%), 32(53.33%), 7(11.67%) for K1, K2, K3 and K4, respectively [Table/Fig-2]. The positive and negative Co-A test is showed in [Table/Fig-3]. Eight (13.33%) of them remained untypable. String test [Table/Fig-4] was found positive among 12 out of 60 (20%) of tested isolates of K.pneumoniae. The occurrence of K1, K2, K3 and K4 serotypes among the hvKP strains of K. pneumoniae was 2(16.6%), 0(0%), 1(8.3%), 1(8.3%), respectively [Table/Fig-5]. Among the isolates studied, 20/60 (33.3%) of K. pneumoniae studied were belonging to ESBL producers [Table/Fig-6]. The drug resistance genes harboring among the capsular serotypes, K1, K2, K3 and K4 of K. pneumoniae in respect to SHV gene was 13(65%), 0(0%), 3(15%) and 1(5%) respectively and for CTX-M-1 it was 12(60%), 0(0%), 5 (25%) and 0 (0%), respectively [Table/Fig-7]. The rate of typability among the serotypes of K. pneumoniae K1 serotype by Co-A was 13(21.6%) and 14(23.3%) by PCR [Table/Fig-8]. It was found to be 0% in case of K2 serotype both by Co-A and PCR. The occurrence of SHV and CTX genes among the K1 serotype in the present study was found to be 65% [Table/Fig-9] and 60% [Table/Fig-10] respectively without the presence of TEM.
Rate of typability among the clinical isolates of K. pneumoniae in respect to various capsular serotypes by Co-A method n=60.
| Serotypes | Rate of Typeability by co agglutination n (%) |
|---|
| K1 | 13 (21.67) |
| K2 | 0 |
| K3 | 32 (53.33) |
| K4 | 7 (11.67) |
Showing positive and negative Co-A test.
Showing positive string test.

Showing hvKP strains of K. pneumoniae harboring K1, K2, K3 and K4 antigens n=12.
| Serotypes | No. of hvKP strains n (%) |
|---|
| K1 | 2 (16.6) |
| K2 | 0 |
| K3 | 1 (8.3) |
| K4 | 1(8.3) |
Results of screening and confirmatory test of ESBL production among 60 isolates of K. pneumonia.
| Screening test positive n (%) | ESBL Positive n (%) |
|---|
| 39 (65) | 20(33.3) |
Showing drug resistance genes among the capsular serotypes of K. pneumoniae (n=20).
| Serotypes | TEM | SHV n (%) | CTX-M-1 n (%) |
|---|
| K1 | 0 | 13 (65) | 12 (60) |
| K2 | 0 | 0 | 0 |
| K3 | 0 | 3 (15) | 5 (25) |
| K4 | 0 | 1 (5) | 0 |
Comparison of serotypes isolated among the isolates of K. pneumoniae by Co-A and PCR n=60.
| Serotypes | Rate of Type ability by Co-agglutination n (%) | Rate of Type ability by PCR n (%) |
|---|
| K1 | 13 (21.6) | 14(23.3) |
| K2 | 0 (%) | 0 (0%) |
Serotypes Rate of Type ability by Co-A n (%) Rate of Type ability by PCR n (%)
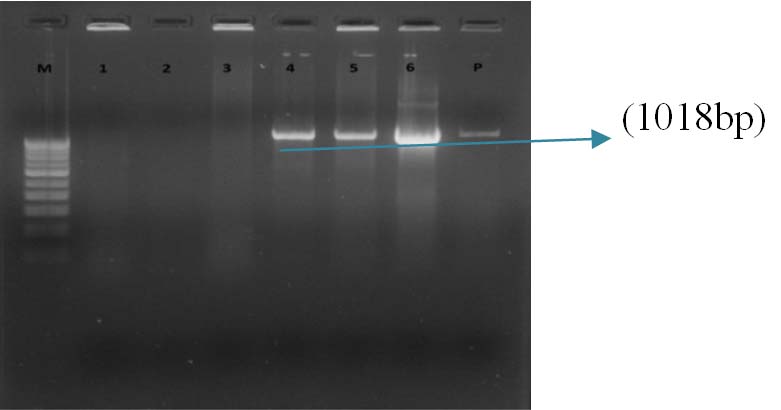
Gel Doc picture showing PCR amplified product of bla SHV. Lane M – 1 Kb DNA ladder, Lane 4-6 bla SHV positive amplicons (1018 bp), Lane 1-3- Isolates of K. pneumoniae without bla SHV, Lane P-Positive control.
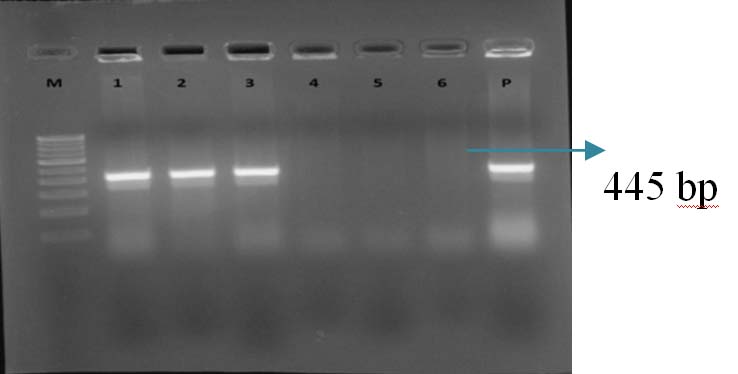
Gel Doc picture showing PCR amplified product of blaCTX-M-1. Lane M – 1 Kb DNA ladder, Lane 1-3 blaCTX-M-1 positive amplicons (445bp), Lane P-Positive control.
Discussion
The present study tried to verify the incompatibility between Co-A method and the PCR method in the identification of the capsular serotypes of K. pneumoniae. Apart from diagnostic value, serotyping has epidemiologic value in differentiating strains within species.
Though PCR diagnosis is highly sensitive and specific, widely used, even detects non-viable organisms, faster, could be performed and interpreted by other than taxonomical expertise, it faces risk of amplicon contamination, unable to detect closely related serotypes, requirement of gel electrophoresis, and the use of health hazardous reagents such as ethidium bromide [21,22].
Although, genotypic methods have higher discriminatory power than phenotypic methods, the PCR technique is expensive and its sensitivity can be restrained by contaminants of DNA extraction solution, concentration of the PCR mixtures, annealing temperature and PCR cycling conditions [23,24].
In this study, the total typable rate was about 86.67% and K1 and K2 serotypes accounted for 21.67% and 0% respectively. Whereas some other study reported that the rate of typability among the isolates of K. pneumoniae was about 77% and K1, K2 serotypes accounted for 21.7% and 9.3% of the tested isolates respectively [25]. The rate of typability in another research was 87.7%. Serotypes K1 and K2 were found to be predominant, accounting for 46.6%, and 20.5%, respectively [26]. At the same time study conducted in Taiwan suggested the occurrence of 18.2% K1 serotype and 16.4% K2 serotype among the isolates of K. pneumoniae [27]. In the present study the rate of typability in both Co-A test and PCR was somewhat similar (Co-A- 21.6% and PCR- 23.3%). Study conducted by Maria EN et al., suggested that, slide agglutination serotyping method showed a better agreement with the PCR results [28]. Another study by LaClaire LL et al., showed an overall 94% agreement between slide agglutination serotyping method and PCR results [29].
The result of string test performed in the present study showed that, out of 60 isolates of K. pneumoniae tested for the occurrence of hvKP strains, only 12 (20%) were showing the positive result. It suggests that most of the isolates studied did not belong to hvKP strains. A study from China showed varied prevalence rate of hvKP among the isolates of K. pneumoniae in different cities. It was found highest percentage of 73.9 and the lowest of 8.3% [30]. Study conducted by Liu YM et al., showed that, 26/70 (37.1%) of K. pneumoniae studied were belonging to ESBL producers, 22(31.4%) were hvKPs. Of the 26 ESBL producers, 2 (7.7%) were found to be hvKP strains [31]. In the study of Lee CH et al., hypermucoviscosity was identified in 35 (38.5%) of 91 K. pneumoniae isolates. Twenty four (26.4%) isolates out of 91 were ESBL producing strains. Only one ESBL producing K. pneumoniae strain expressed the hypermucoviscosity [32]. The study conducted by Gharrah MM et al., suggested a negative association between hypermucoviscosity and ESBL production among the isolates of K. pneumoniae. The result of the same study showed that, 62% of non-ESBLs exhibited hypermucoviscosity compared to the ESBLs (4%) [33]. The present study showed, 20/60 (33.3%) of K. pneumoniae studied were belonging to ESBL producers. Of the 60 serotypes, 12(20%) were hvKPs. Out of 20 ESBLs 4 (20%) were hvKPs. The present study showed the occurrence of 2(16.6%) of K1 serotype, 0% of K2 serotype, 1(8.3%) of K3 serotype and 1(8.3%) of K4 serotype among the hvKP isolates studied. But the study conducted by other researcher showed 23.8% of K1 serotype and 42.9% of K2 serotype among the hvKP isolates of K. pneumoniae [34]. At the same time, the study conducted by Qu T et al., also showed high prevalence rate of K1 serotype (68.9%) and the occurrence of K2 serotype (20%). The study also showed that all the hvKP strains belonged to K1/K2 serotype [35]. In another study, the occurrence of K1, K2 and non K1/K2 serotypes ranged from 47-81%, 20% and 23-33% respectively [36]. Study conducted by Cheong HS et al., suggested that, of the 120 K. pneumoniae isolates, 20 (16.7%) were found to produce ESBLs and among the ESBL-producers, only three were proved to be serotype K1 [37]. Whereas our study showed, out of 60 isolates studied, 39 (65%) isolates were showing the resistance to third generation cephalosporin and out of 39 screened resistant isolates, 20(51.28%) was proved to produce ESBLs and among the ESBL producing K. pneumoniae isolates, 9 (45%) isolates were proved to be K1 serotype.
In the present study, the occurrence of SHV and CTX genes among the K1 serotype was found to be 65% and 60% respectively. None of the capsular serotypes studied were showing the genes coding for blaTEM. But the study conducted by Zang Y et al., suggested that, the isolated serotypes were mostly belonging to K1 serotype and it was found to harbor blaTEM, blaSHV and blaCTX-M genes [30]. Hence the study well established that capsular serotypes are not only responsible for virulence but also for drug resistance.
Limitation
Co-A and PCR study of serotypes other than K1, K2, K3 and K4 would have added more significance to the study.
Conclusion
Co-A test is a good method of serotyping as it is cheap, sensitive, and quick in diagnosis, processed without any requirement of expensive equipments and experts to handle the test. So, the test can be used not only for diagnostic purpose but also for epidemiological study. The highest number of hvKP strains among the tested isolates was harboring K1 serotype. Most of the K1 capsular serotypes of tested K. pneumoniae strains were exhibiting the genes coding for both SHV and CTX genes. The study suggested that hvKP strains are not only responsible for virulence but also for drug resistance.
Serotypes Rate of Type ability by Co-A n (%) Rate of Type ability by PCR n (%)