Bloodstream infections are a serious cause of morbidity and mortality amidst healthcare settings. Bacteraemia due to gram-negative bacilli is a huge challenge to the clinicians because of the rapidly emerging multidrug resistance among these microorganisms [1]. The mortality reported as a cause of Gram negative bacteraemia ranges from 12-38% [2]. The prime risk factors associated with Gram negative bacteraemia include broad spectrum antibiotic treatment, immunosuppressive therapy, burns/trauma, anatomic obstruction, extremes of age, malignancy, diabetes, pulmonary disease, chronic haemodialysis, HIV infection, presence of intravenous and central venous catheters. Infections of the respiratory tract, urinary tract, gastrointestinal tract, hepato-biliary tract, and skin or soft tissues are the common sources of bloodstream infections [3].
Gram negative bacteria associated with bacteraemia differ by the onset of the infection (hospital or community) and also by the primary source of infection. Escherichia coli, Klebsiella pneumoniae, Psuedomonas spp. and Enterobacter spp. are among the chief nosocomial pathogens that have been reported to be associated with sepsis whereas species of Salmonella are associated with the community acquired bacteraemia [4].
Antimicrobial resistance among the gram negative bacilli is increasingly rising, especially against β-lactams due to the production of β-lactamase enzymes by these organisms. Types of β-lactamases confering resistance to expanded-spectrum cephalosporins include chromosomally mediated β-lactamases, Extended-Spectrum β-Lactamases (ESBLs), Metallo-β-lactamases, Klebsiella Pneumoniae Carbapenemases (KPCs), and the New Delhi Metallo-β-lactamases (NDM). Previously done studies have reported the highest ESBL rates in K. pneumoniae from Indian subcontinent (72%). The prevalence of Amp C β-lactamases in blood culture isolates of E. coli and Klebsiella spp. is reported as 8-43%. The prevalence of carbapenemases in Klebsiella pneumoniae, E.coli is reported as 51% and 28% respectively. Among these carbapenemase producers, 79% of Klebsiella strains and 28% of E. coli were Metallo-β-lactamases (MBL) producers. The remaining isolates could be KPC/OXA-48/VIM/IMP producers or Amp C β-lactamases producers+Porin loss [5].
The current study is undertaken to note down the clinical and microbiological aspects of bacteraemia caused by coliforms among adults with special reference to antimicrobial resistance among these pathogens and the detection of the enzymes and genes by phenotypic and genotypic methods. The risk factor analysis and antibiotic audit will help us to look into the areas of interventions which can be taken up to decrease the morbidity as well as mortality. We also aim to look into the outcome among the bacteraemic patients.
Materials and Methods
This hospital based descriptive study was carried out over a period of 18 months from October 2015 to April 2017 in the Microbiology laboratory of a tertiary care centre. The study included 175 blood culture samples of the adult patients admitted with clinically suspected sepsis, in whom, blood culture yielded Coliforms. The samples of paediatric age group, patients with negative blood culture or with positive blood culture yielding gram negative bacilli other than Coliforms, gram positive cocci and yeasts were excluded.
The study was approved by the Institutional Ethics Committee. The data was collected using a proforma and medical records were examined for information like age, gender, duration of hospital stay, stay in the ICU, history of chronic disease, underlying infections, history of instrumentation or device implantation i.e., (the risk factors and co-morbidities) any surgical procedure performed, other investigations done and the treatment given. Bacteriological culture from other sites was noted to trace the probable source of bacteraemia.
Chi square test was done to do the comparison of different risk factors in cases of coliform bacteraemia and control group with patients having bacteraemia other than coliform bacilli. Odds ratio was also estimated to find out the association between the various risk factors. Statistical significance was considered when p was less than 0.05. The statistical package SPSS version 17.0 was used to perform the analyses.
Microbiological Methods
Blood culture was done by automated blood culture system (BacT/ALERT; bioMeriux, Inc. Durham, North California, USA). A gram stained smear preparation was done from the broth of positive blood culture bottles. The positive blood culture bottles were sub-cultured on Blood agar, Chocolate agar, and MacConkey agar and incubated aerobically overnight at 370C. The cultured plates were examined after 24 hours and organisms identified by their colonial morphology, Gram staining and appropriate conventional biochemical tests and Vitek 2 Compact system (bioMeriux, Inc. Durham, USA).
The Antibiotic Susceptibility Testing (AST) was carried out using Vitek Compact 2 system and Modified Kirby Bauer disc diffusion method. The antibiotic discs (Himedia) used for coliforms included – Ampicillin (10 mcg), Amoxicillin/Clavulanic Acid (30 mcg), Cefuroxime (30 mcg), Ceftriaxzone (30 mcg), Cefepime (30 mcg), Cefoperazone/Sulbactam (75/30 mcg), Amikacin (30 mcg), Gentamycin (10 mcg), Ciprofloxacin (5 mcg), Imipenem (10 mcg), Meropenem (10 mcg), Piperacillin/Tazobactam (100/10 mcg), Colistin (10 mcg). The interpretation of results was based on the recommendations of Clinical Laboratory Standards Institute (CLSI). ESBL production in Klebsiella spp. and E.coli was detected by the screening and confirmatory tests recommended by CLSI guidelines [6].
The isolates of E.coli, Klebsiella spp. with MIC of 2-4 μg/ml for Imipenem/Meropenem or the isolates which were resistant to Ertapenem in disk diffusion were tested further for presence of carbapenemases using the MHT as per the CLSI guidelines [6].
MHT positive strains of E.coli, Klebsiella spp. were further tested for MBL production by using Imipenem with and without EDTA Ezy MIC strip (Himedia) [7].
The cefoxitin-cloxacillin disk diffusion test was performed as described in the previous study [8].
Molecular Detection of the blaNDM 1 and blaKPC gene by Conventional Polymerase Chain Reaction (PCR)
The DNA was extracted according to CDC protocol by boiling method, from freshly cultured well isolated bacterial colonies grown on 5% sheep blood agar plates following overnight incubation. The primer sets were purchased from Sigma chemicals (Sigma Aldrich Pvt., Ltd.,) and the sequence was:
Primer set blaNDM-1
NDM1 - Forward (5’-CACCTCATGTTTGAATTCGCC-3’) 561 base pair
NDM1 - Reverse (5’-CTCTGTCACATCGAAATCGC-3’) 561 base pair
Primer set blaKPC
KPC - Forward (5’-CATTCAAGGGCTTTCTTGCTGC-3’) 538 base pair
KPC - Reverse (5’-ACGACGGCATAGTCATTTGC-3’) 538 base pair [9]
PCR conditions: The PCR cycle was run at the following temperature and time settings: 94°C for 10 minutes for initial step of denaturation, followed by 30 cycles of 40 seconds at 94°C, 40 seconds at 55°C and 1 minute at 72°C and lastly a final extension step of 7 minutes at 72°C.
DNA extracted from bacterial strains of K. pneumoniae ATCC BAA-1705 and E. coli ATCC 25922 were also included in the testing panel as positive and negative control strains respectively in each PCR. The resulting PCR products along with the DNA ladder were loaded in wells of 1% agarose gel prepared in 1X Tris EDTA (TAE) buffer. The DNA fragments were segregated by gel electrophoresis at 120 V for 45 minutes. The gel was then stained with Ethidium bromide for 30 minutes and visualized under UV light. A photograph of the gel was taken by Alpha Imager software.
Results
A total number of 5385 blood samples were obtained in 12 months duration of sample collection i.e., from Oct 2015-Oct 2016. Out of 5385 blood samples received, 700 (12.9%) showed a positive growth by automated system BacT/ALERT. The Coliform bacilli including Escherichia coli, Klebsiella spp, Enterobacter spp. and Citrobacter spp. constituted 175 of 700 (25%) isolates. E. coli was the major pathogen isolated among Coliforms. The distribution of coliforms is shown in [Table/Fig-1]. Maximum patients (38/175) were from the age group 58-67 years and broadly most patients (143/175) belonged to 38 years to 77 years of age. A male preponderance (58.1%) was noted among the bacteraemic patients.
Distribution of coliform bacilli in blood culture.
| Isolate | Number (%) |
|---|
| Escherichia coli | 101 (57.71) |
| Klebsiella spp. | 60 (34.28) |
| Enterobacter spp. | 12 (7.42) |
| Citrobacter spp. | 2 (1.7) |
| Total | 175 |
Time to positivity (in hours) of the coliform isolates ranged from 5-36 hours. The maximum positives were in 12 hours. The range of mean time to positivity of isolates with maximum positives was 9-18 hours.
The common infections associated with coliform bacteraemia were UTI (33.1%), pneumonia (18.3%), gastroenteritis (8.6%), organ abscess (6.8%), diabetic foot (6.3%), cellulitis (5.7%) osteomyelitis (1.7%). Source of bacteraemia is depicted in [Table/Fig-2]. The statistically significant risk factors included previous hospital stay, invasive procedures, ICU stay, previous antibiotic use, previous surgery, respiratory and renal disease, diabetes and hypertension. (p-value<0.001)
Source of infection in coliform bacteraemia.
| Source of Bacteraemia | Nuber of patients (n) | Percentage (%) |
|---|
| Genitourinary tract | 58 | 33.1% |
| Skin and soft tissue | 41 | 23.4% |
| Intra venous catheter/ Chemoport site/ Urinary catheter/ Central venous catheter | 38 | 21.7% |
| Lungs | 32 | 18.2% |
| Gastrointestinal tract | 25 | 14.2% |
| Untraced | 17 | 9.7% |
| Hepatobiliary system (Gall bladder, Pancreas) | 12 | 6.8% |
| Bone | 3 | 1.7% |
| Muscle | 5 | 2.8% |
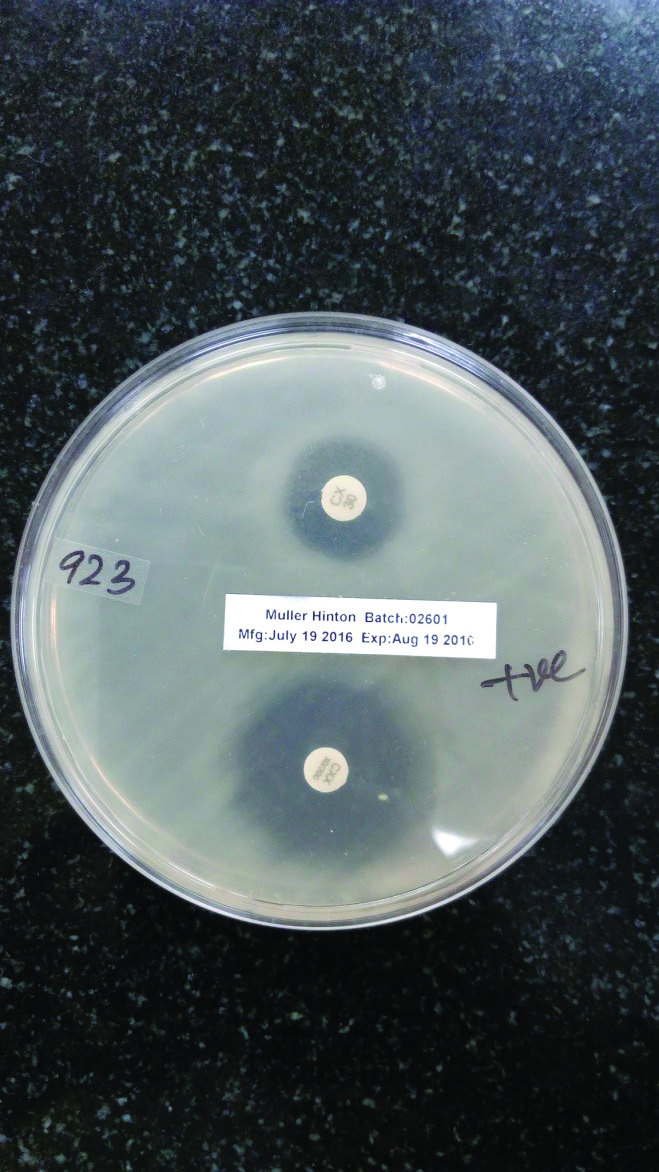
Antibiotic resistance pattern is shown in [Table/Fig-3]. The rate of ESBL enzyme production in E.coli and Klebsiella spp was 44.7% with a rate of 42.5% in E.coli and 48.3% in Klebsiella spp. respectively. Out of total 161 isolates, a total of 68 (42.2%) isolates were resistant to Cefoxitin after the screening test. These 68 cefoxitin resistant isolates were further confirmed for Amp C production by performing Cefoxitin-Cloxacillin disk diffusion test, 35 (51.4%) isolates were positive with Cefoxitin-Cloxacillin test of which 26 were E. coli and 9 were Klebsiella spp.
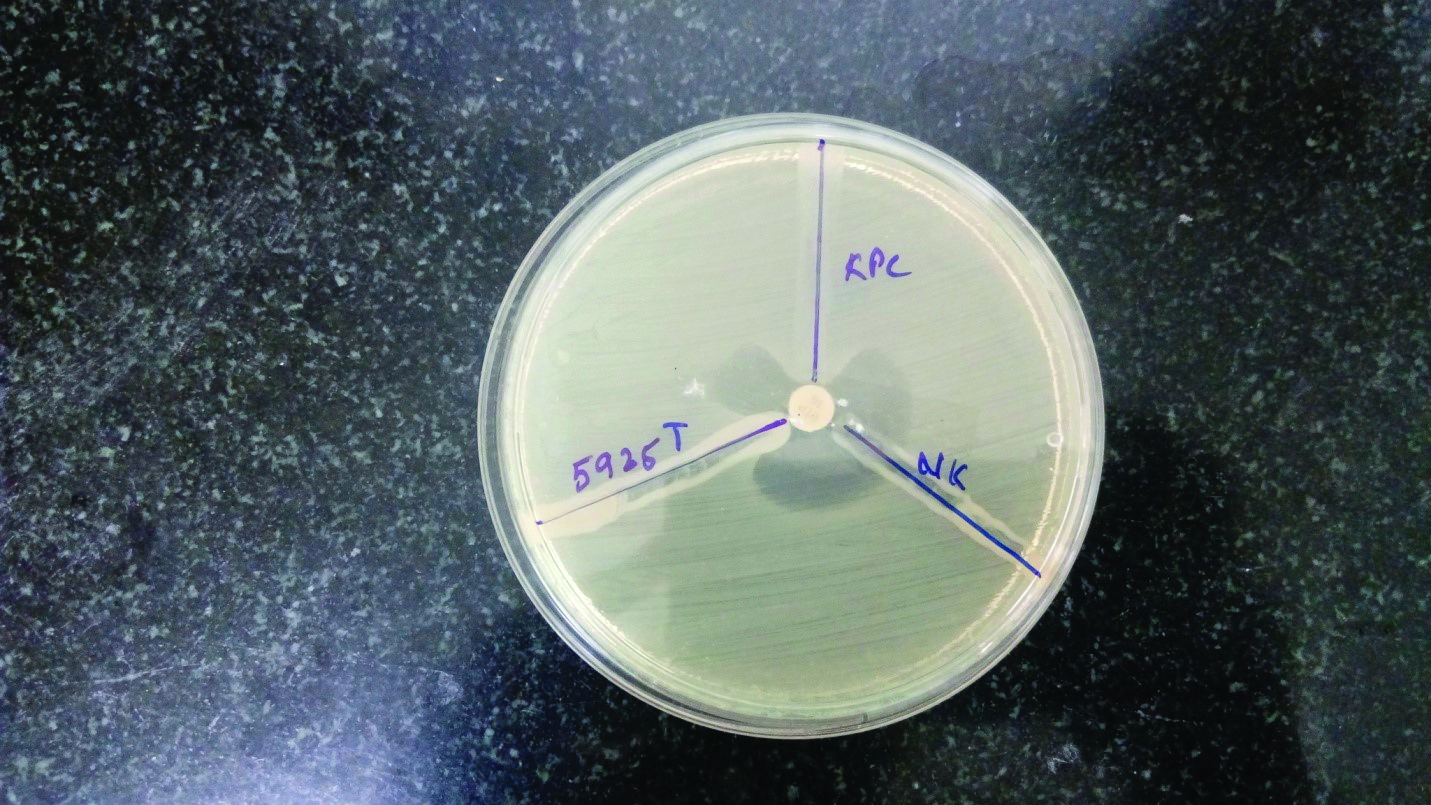
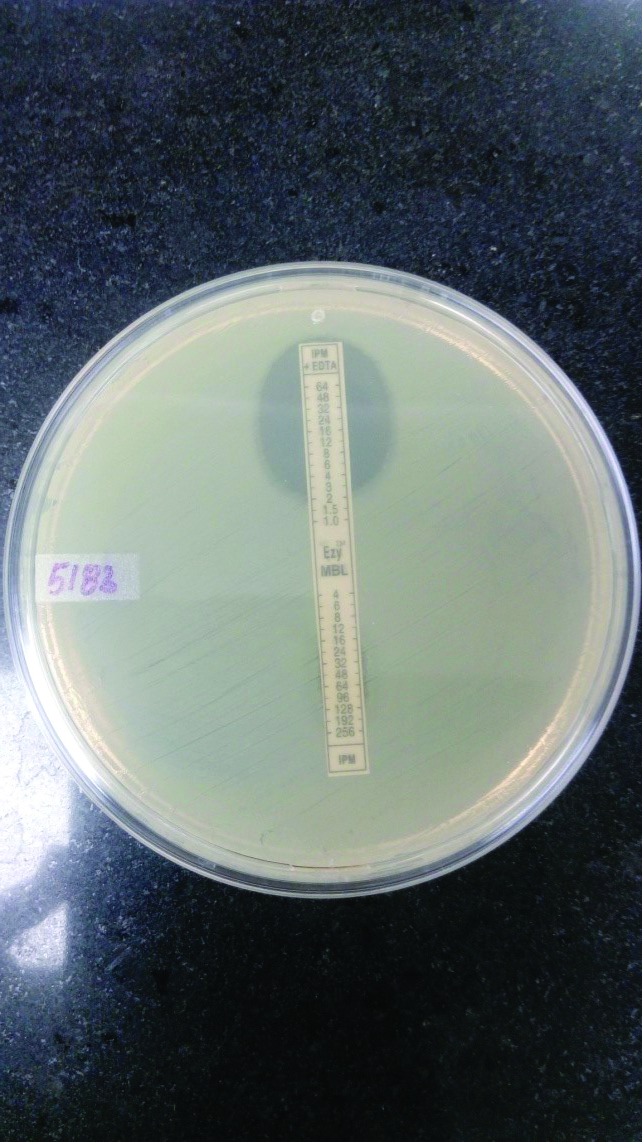
Screening test for detection of Carbapenem resistance was performed on 175 Coliform isolates and 25 isolates were found to be resistant (14.2%) [Table/Fig-4]. These 25 isolates were subjected to MHT to confirm the Carbapenem resistance of which 11 were positive (7:Klebsiella spp., 4:E. coli.). The 25 Carbapenem resistant isolates were also tested for MBL production by E-test. 14, out of the 25 isolates were MBL producers (10:Klebsiella spp. .4:E. coli).Genotypic results are shown in [Table/Fig-5]. The AmpC, MHT, MBL detection and PCR results are shown in [Table/Fig-6,7,8 and 9].
Antibiotic resistance pattern of the coliforms (n=175).
| Antibiotic | E. coli (Total=101) | Klebsiella spp. (Total=60) | Enterobacter spp. (Total=12) | Citrobacter spp. (Total=2) |
|---|
| Ampicillin | 77.2% (n=78) | 75% (n=45) | 33.3% (n=4) | 100% (n=2) |
| Amoxicillin/Clavulanic acid | 33.6% (n=34) | 28.3% (n=17) | 66.6% (n=8) | 100% (n=2) |
| Cefuroxime | 44.5% (n=45) | 43.3% (n=26) | 50% (n=6) | 50% (n=1) |
| Ceftriaxone | 46.5% (n=47) | 40% (n=24) | 41.6% (n=5) | 100% (n=2) |
| Cefepime | 25.7% (n=26) | 30% (n=18) | 25% (n=3) | 100% (n=2) |
| Cefoperazone/ Sulbactam | 15.8% (n=16) | 15% (n=9) | 0% (n=0) | 50% (n=1) |
| Amikacin | 21.7% (n=22) | 25% (n=15) | 8.3% (n=1) | 50% (n=1) |
| Gentamycin | 32.6% (n=33) | 30% (n=18) | 33.3% (n=4) | 100% (n=2) |
| Ciprofloxacin | 66.3% (n=67) | 63.3% (n=38) | 8.3% (n=1) | 100% (n=2) |
| Imipenem | 7.9% (n=8) | 25% (n=15) | 8.3% (n=1) | 50% (n=1) |
| Meropenem | 7.9% (n=8) | 25% (n=15) | 8.3% (n=1) | 50% (n=1) |
| Piperacillin/Tazobactam | 12.8% (n=13) | 18.3% (n=11) | 0% (n=0) | 100% (n=2) |
| Colistin | 0% (n=0) | 1.6% (n=1) | 25% (n=3) | 0% (n=0) |
Results of the phenotypic tests done for detection of carbapenemase, metallo-beta-lactamase.
| Phenotypic Test results | Total isolates | E.coli | Klebsiella spp. |
|---|
| MHT positive isolates (Carbapenemase producers) | 11/25 (44%) | 4/11 (36.3%) | 7/11 (63.6%) |
| E-test positive isolates (MBL producers) | 14/25 (56%) | 4/14 (28.5%) | 10/14 (71.4%) |
Results of conventional PCR for detection of blaNDM-1 and blaKPC gene.
| Organism | blaNDM-1 | blaKPC |
|---|
| E. coli | 6 (37.5%) | 0 |
| Klebsiella spp. | 10 (62.5%) | 0 |
Phenotypic detection of AmpC beta lactamase production by Cefoxitin-Cloxacillin disk diffusion test.
Top: Cefoxitin disk
Bottom: Cefoxitin-Cloxacillin disk
Phenotypic detection of carbapenemases by Modified Hodge test.
Phenotypic detection of MBL production by E-test.
Top: Imipenem with EDTA, MIC= 2μg/ml,
Bottom: Imipenem No zone
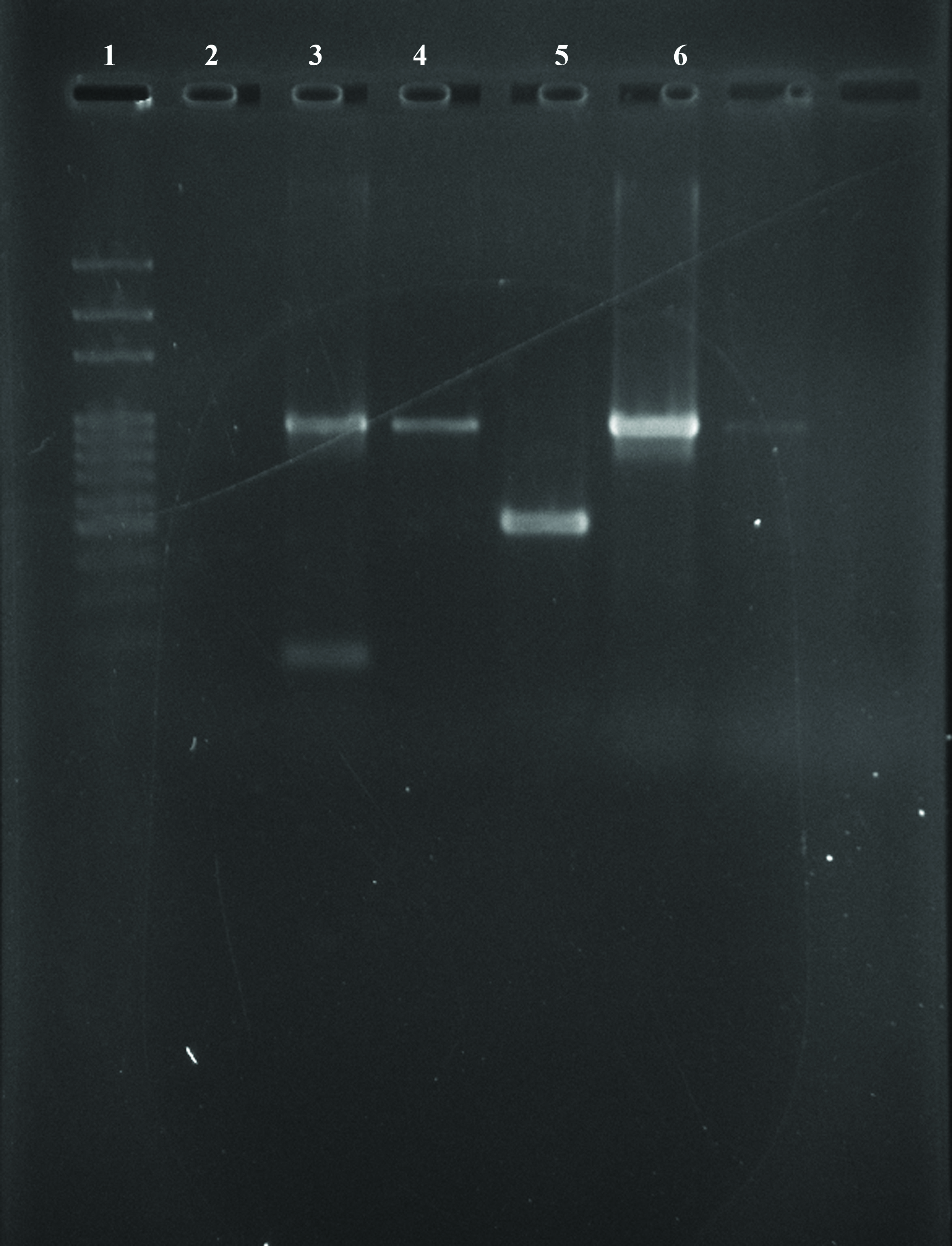
Agarose gel picture of amplified products showing blaNDM-1 gene.
Lane 1: DNA ladder (100 bp)
Lane 2: Sample 1- Negative for blaNDM-1 and blaKPC gene
Lane 3: Sample 2 – Positive for blaNDM-1 gene
Lane 4: Sample 3 – Positive for blaNDM-1 gene
Lane 5: Positive control for blaKPC gene
Lane 6: Positive control for blaNDM-1 gene
Discussion
Bloodstream infections appear to be the chief reason of morbidity and mortality. In the recent few years, the Coliform bacilli have emerged as alarming pathogens in causing bacteraemia and leading to sepsis and further septic shock resulting in fatal consequences among the adult patients.
Our study revealed that the Coliform bacilli constituted 25% of the total positive blood cultures. Among the Coliforms, E. coli 101 (57.7%) was the supreme organism to cause bacteraemia in adults followed by Klebsiella spp. 60 (34.2%) in our area. Study by Abhilash KP et al., showed 102 (77.86%) episodes of bacteraemia were caused by E. coli and 29 by (22.14%) Klebsiella spp [10].
The Time To Positivity (TTP) is considered as a representative marker of the magnitude of bacteraemia and a handy tool for diagnosis and prognosis in bacteraemic cases. The investigation done in present study shows that the maximum proportion of E.coli and Klebsiella spp. isolates were positive by 12 hours in the automated blood culture systems. The range in hours for most of the pathogens to be positive was 9-18 hours.
The statistics of current study unveil that UTI was the most frequent infection associated with bacteraemia leading to urosepsis as a complication in 33.1% of the cases. followed by skin and soft tissue as a focus of infection in 41 (23.4%) cases, Intra venous catheter/ Chemoport site/ Urinary catheter/ Central venous catheter in 38 (21.7%) patients, lungs leading to pneumonia in 32 (18.28%) and focus of infection from the gastrointestinal tract including the peritoneum in 25 (14.2%) cases. This data was synchronous with the findings of Abhilash KP et al., [10].
In 2011, HV Prashanth and others concluded in their study done in South India that the most common predisposing factors were diabetes, previous surgery, malignancies, burns, HIV/AIDS, cirrhosis and neutropenia [5].
The antibiogram of the isolates in our study revealed that overall the isolates were most resistant to Ampicillin and most sensitive to Colisitn. Most Coliform bacilli showed high resistance to Penicillins, Fluoroquinolones, 3rd generation cephalosporins, Beta lactamase inhibitor combination and Aminoglycosides. The antibiotic resistance rates in a study performed by Gupta S and fellows during 2016 in Northern part of India were amoxiclav (91.4%), cefixime (90.37%), ceftriaxone (74.7%) and cefoperazone/sulbactam (81.14%) [11].
In our hospital the rate of ESBL producing Klebsiella spp. was 48.3% which was more compared to the ESBL producing E.coli, 42.5%. In a study conducted in North India by Pathak A et al., the ESBL rates were 69% for E. coli and 41% for K. pneumonia [12].
AmpC beta lactamase producers in our study were 42.2%; while comparing with other studies we found that a rate of 37.50% has been reported from Chennai by Subha A et al., [13].
The incidence of carbapenem-resistant Gram-negative rods is on the rise all along the globe, posing a huge public health threat. In the present study total resistance to Carbapenems in E.coli and Klebsiella spp was high accounting for 14.28%. The data of earlier studies done reveal that Klebsiella spp. and E. coli account for the largest proportion of CRE, namely 39.3% and 22.0%, in a study done by Yanling XU et al., [14]. All the 25 isolates that showed resistance to Carabapenems in screening test were subjected to MHT and we found that 44% (11 out of 25) of the isolates were picked up as positive by MHT while the remaining 14 isolates were negative. A study done by Solanki R et al., found 52 out of 100 carbapenem resistant Gram negative isolates were picked by MHT [15]. CLSI recommends MHT as the only method to detect Carbapenemase production since it is highly sensitive and specific method to detect the KPC type of Carbapenemases. Although, the strains that produce excessive amount of Amp C beta-lactamases and CTX-M type of ESBLs may give rise to false positives. Conversely, isolates that produce Metallo-beta-lactamases and New Delhi Metallo-beta-lactamases or any other enzymes that express feeble Carbapenemase activity can lead to false negative results. By and large, the sensitivity and specificity of MHT to detect the presence of MBL and NDM is low, being 77% for MBL and 39% for NDM respectively [15]. A 56% of our isolates were MBL producers by E- test using Imipenem with and without EDTA Ezy MIC strip (Himedia).
PCR: Out of these 16 strains with blaNDM, 10 were Klebsiella spp. and the remaining six were E.coli. However, we could not find any isolate harbouring blaKPC gene. In a study performed by Solanki et al., at Hyderabad, out of 100 carbapenem resistant isolates, 37 were having blaNDM-1gene. On the other hand they found only 1 blaKPC gene which was an isolate of E.coli. This was synchronous with our results showing that the Klebsiella spp. harboring blaNDM-1gene outclassed the E.coli with blaNDM-1gene. Also, the rate of isolation of blaKPCgene was extremely low or nil in both the studies [15]. The above findings imply that the out of the total 25 isolates resistant to Carbapenemases, 16 were harboring the blaNDM-1(NDM producers) however, the remaining 9 isolates could be having other mechanisms of Carbapenem resistance like production of OXA,VIM,IMP, hyperproduction of AmpC and ESBL, porin loss and efflux pump mediated resistance.
Antimicrobial therapy remains the mainstay of treatment for sepsis, along with the most favourable management of its consequences (like, shock or metastatic suppurative complications), and surgical intervention to treat an infection site (e.g., debridement, abscess drainage, or removal of intravascular devices), when apt.
The combination therapy with two or more antibiotics with different mechanism of action like betalactam with aminoglycosides, fluoroquinolones or macrolides administered within 6 hours of sepsis presentation is recommended. Carbapenems are the preferred drugs among the beta lactams. Kang and fellow researchers found that carbapenem and ciprofloxacin were the most efficient antibiotics in the treatment of ESBL bacteraemia by E.coli and Klebsiella spp. in their study conducted in Korea [16].
Clinical outcome: In Kang’s study the mortality rate was 25.6% i.e., 34 of 133 cases could not survive [16]. Fortunately, the total mortality rate due to coliform bacteraemia in our study was not very high; accounting 11.6%.
Conclusion
Further studies are required to determine the epidemiology of coliform related infections, their mechanisms of resistance, antimicrobial susceptibility pattern and the associated clinical burden locally. Prospective studies into transmission, molecular characterization, and, most importantly, treatment regimens are desperately needed as top priority. Moreover, the invention of new antimicrobial agents and non-traditional antimicrobial methods should have a prime concern globally.