OSCC is the sixth most common cancer in the world and is of significant public health importance, especially in the developing countries. According to Globocan 2012 data, 600,000 new cases and 255,000 deaths worldwide and 160,000 new cases and 77,000 deaths in India were reported [1].
HPV are a group of large family of non-enveloped DNA viruses, mainly associated with cervical cancers [2-4]. More than 100 subtypes of HPV exist. While most of them present asymptomatically and not treated, some strains may cause harmless warts and nearly 40 strains can easily spread through direct sexual contact [5]. Sexually transmitted HPV can be categorised as: (a) High risk; and (b) Low risk. Low risk HPV subtypes do not cause cancer but causes warts on and around genitals, mouth or throat. The subtypes involved in the cancer development are high risk subtypes, particularly HPV-16, 18, 31, 33 and 45 [6]. The oncogenic proteins of high risk HPV such as E6 and E7 are capable of inducing cellular transformation by degrading cell cycle regulators p53 and Rb leading to immortalization of cells [7,8]. Recent epidemiologic studies have suggested that HPV may be an independent risk-factor for oropharyngeal and anogenital cancers [9-11]. So in 2012, International Agency for Research on Cancer (IARC) included HPV infection as one of the risk-factors for the development of OSCC. Evidence now suggests HPV may not only modulate the malignancy process in some tobacco- and alcohol-induced oropharynx tumours, but might also be the primary oncogenic factor for inducing carcinogenesis among some non-smokers [12].
There are only a handful of studies attributing HPV infection in OSCC in the Indian context. Therefore, more evidence is needed regarding oral HPV prevalence among OSCC subjects to estimate the risk.
Absence of well-defined early symptoms is one of the major factors in lack of diagnosis for OSCC not only in India but also elsewhere. Saliva serves as a non-invasive tool in the clinical diagnosis of patients’ with oral cavity diseases. Many biomarkers have been identified in saliva, which reflects the environmental status in and around the oral cavity, especially in cancers of the head and neck [13-15]. Detection of HPV in the saliva of OSCC or high risk subjects could serve as a screening module that could be applied clinically to a large population. So, the present study was aimed at detecting and genotyping HPV in the saliva of OSCC subjects, subjects who are frequently exposed to risk factors leading to OSCC (high risk subjects) and normal subjects by nested PCR using capsid protein L1 specific primer.
Materials and Methods
This was a prospective, cross-sectional study carried out from October 2016 to August 2017 involving JSS Dental College Hospital and Bharath Hospital & Institute of Oncology, Mysuru, Karnataka, India. The samples were analysed in the Centre of Excellence of Molecular Biology & Regenerative Medicine, Department of Biochemistry, JSS Medical College Hospital, Mysuru, Karnataka, India.
Study Groups: Sample collection was carried out according to the protocol approved by the Institutional Ethical Committee and informed written consent was obtained. Subjects were divided into three catogories viz., OSCC (histopathologically confirmed), high risk (people who were healthy but had habit of chewing betelnut, smoking and alcohol consumption for atleast 10 years) and normal healthy individuals. Thirty subjects were recruited under each group. Group 1 were normal subjects (mean age 47 years and male:female ratio 18:12), group 2 were high risk subjects (mean age 42 years and male:female ratio 18:12) and group 3 were OSCC subjects (mean age 56 years and male:female ratio 17:13). Clinical data and information related to lifestyle were obtained from clinical charts and personal communication.
Saliva Collection: All the study participants were requested to refrain from consuming any food or beverage 1 hour prior to the saliva collection. At the time of collection, the subjects were given 10mL of 0.9% normal saline and were asked to gargle within the mouth thoroughly. After rinsing the mouth thoroughly, the subjects were asked to expectorate the entire contents of their mouth into a sterile labelled container. All the samples were immediately stored at -200C freezer until further processing.
DNA Extraction: DNA from the saliva samples was extracted using QIAamp DNA mini isolation kit (QIAGEN, Germany) according to the manufacturer’s instructions with minor modifications. Accordingly, the saliva samples were centrifuged at 2000rpm and the pellet was washed in 200μL of 1X PBS and transferred into a 2mL Eppendorf tube. After removing the PBS completely, 200μL of lysis buffer and 20μL of Proteinase K was added to the pellet and was briefly vortexed for 20 seconds. This mixture was then incubated at 560C for 10 minutes. Two hundred microliters of absolute ethanol was added with mixing and the entire lysate was transferred into the spin column. The spin columns were centrifuged at 10,000rpm, 40C for 10 minutes. The collection flow-through was discarded. Then 500μL of pre-wash buffer was added to the column and centrifuged at 10000rpm for 1 minute. The flow-through was discarded and the procedure was repeated again. The spin columns were placed into a sterile collection tube. Approximately, 70-100μL of elution buffer was added and incubated for 5 minutes at room temperature. The DNA was quantified using a Nanodrop (DeNovix) and to determine the quality of the DNA, each DNA was tested by electrophoresis on 2% agarose gels stained with ethidium bromide. The extracted DNA was stored at -200C until further processing for PCR.
HPV Detection by Nested Polymerase Chain Reaction (Nested PCR)
Following the DNA extraction, a nested PCR was performed to detect the presence of HPV in the samples. Oligonucleotide primers (Sigma) were synthesised commercially and primary PCR was performed using a pair of oligonucleotide primers, PGMY09/PGMY11 that targets a 450bp region in L1 gene. A brief master mix containing 2U/μL Taq polymerase, 200nM of dNTPs, buffer with 2.5mM MgCl2 and 200nM of each forward (PGMY09) and reverse (PGMY11) primers were prepared. To this master mix, 50-100ng of DNA template was added and the reaction was made up to 30μL using PCR grade water. Amplification was performed as described elsewhere [16]. Beta-globin gene was used as internal control and DNA from HPV positive cell lines SiHa (HPV 16 +ve) and HeLa (HPV 18 +ve) were used as positive control. For the nested PCR, primarily amplified PCR product was used as template with the same PCR conditions except the primers used were GP5+/GP6+ that targets 150bp region. The positive DNA samples from the nested PCR was then employed in the type specific PCR with appropriate oligonucleotide primers to genotype the most common high risk subtypes such as HPV-16 and HPV-18. The amplified PCR products were elecrophoresed on 2% agarose gel with a 100bp ladder (Himedia). The following were the primer sequences used: PGMY09/PGMY11: CGTCCMARRGGAWACTGATC/ GCMCAGGGWCATAAYAATGG, GP5+/GP6+: TTTGTTACTGTGGTAGATACTAC/ GAAAAATAAACTGTAAATCATAT, HPV16: CACAGTTATGCACAGAGCTGC/ CATATATTCATGCAATGTAGGTGTA and HPV18: CACTTCACTGCAAGACATAGA/ GTTGTGAAATCGTCGTTTTTCA.
Throughout the PCR processing, the recommended stringent precautions were followed and the results were evaluated and compared with the use of appropriate positive and negative controls, to avoid cross-contamination and false positive reactions.
Statistical Analysis
All the data were tabulated in Microsoft Excel spreadsheet and were expressed in mean±SD or in percentages. Chi square test was performed to test the association between the categorical variables. Odds ratio was calculated using MEDCALC statistical software (https://www.medcalc.org/calc/odds_ratio.php). A p-value of less than 0.05 was considered to be statistically significant.
Results
The results of the present study are based on the observations made on 30 OSCC subjects, 30 high risk subjects exposed to the risk-factors and 30 normal healthy subjects. DNA was extracted from all the samples and were subjected to nested PCR to detect and genotype HPV subtypes. The integrity of all the DNA samples and the adequacy of the PCR program were analysed using β-globin gene and as expected 80% of the extracted DNA had intact β-globin gene which was amplified perfectly resulting in a product length of 409bp. A pair of consensus primers, PGMY09/PGMY11 and GP5+/GP6+ primers was used to detect the presence of HPV and with an optimised nested PCR both the primers gave us a positive result [Table/Fig-1,2]. Ultimately all the positive samples were genotyped with type specific PCR using primers for high risk HPV-16 and HPV-18.
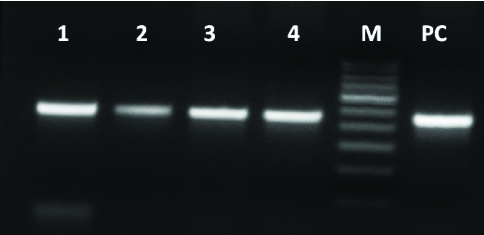
Detection of HPV using consensus primer PGMY09/PGMY11 targeting 450bp L1 region. (Lane 1-4= Positive samples, M= 100bp marker, PC= Positive control).
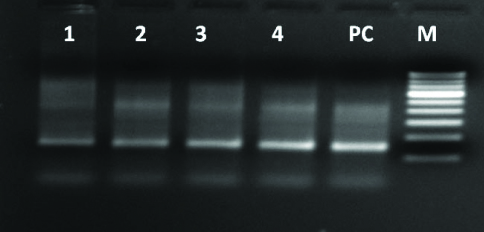
Nested PCR using GP5+/GP6+ primers amplifying a 150bp region with products of primary PCR as template. (Lane 1-4=Positive samples, PC= Positive control, M= 100bp marker).
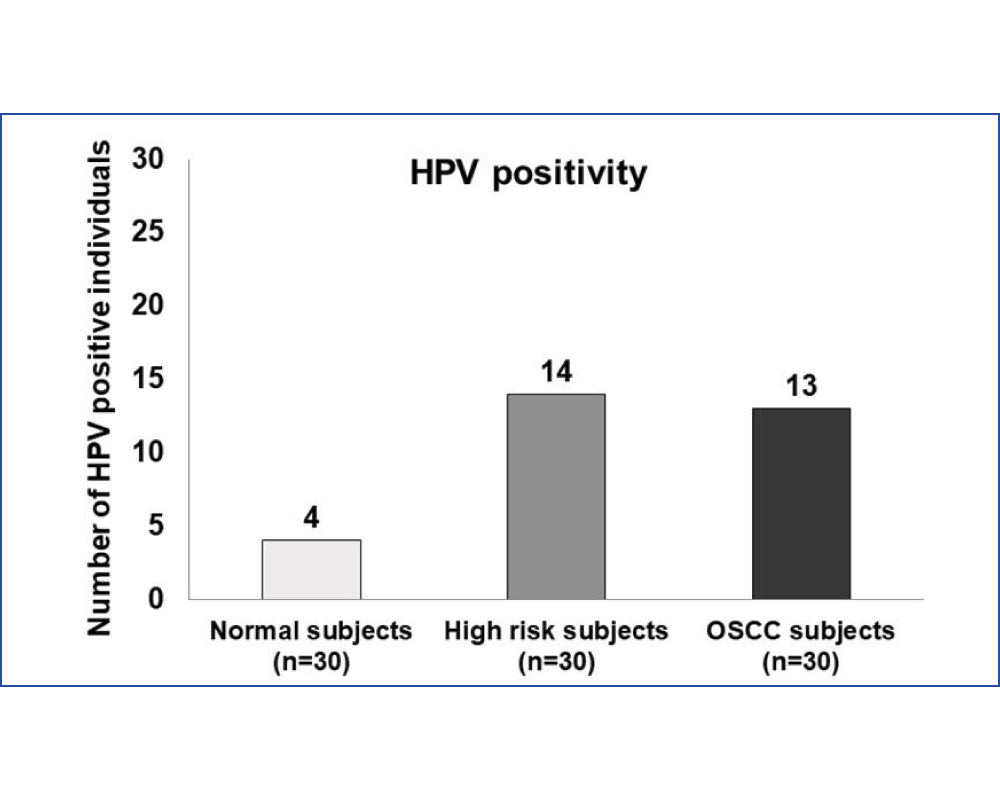
In the present study, HPV was detected in all the groups. A total of 4 (13.3%) normal subjects, 14 (46.6%) high risk subjects and 13 (43.3%) OSCC subjects were HPV positive [Table/Fig-3]. In all the three groups the prevalence of HPV 16 was significantly higher than HPV 18. The odds ratio for high risk subjects to be HPV positive was found to be 5.68 (p= 0.007*) and for OSCC subjects to be HPV positive was 4.97 (p= 0.01*).
HPV status in the study groups.
Association of HPV positivity to risk-factors among high risk and OSCC groups showed that majority of the population who had the habit of smoking and consuming alcohol were HPV positive (61.5% and 63.6% in high risk and OSCC groups respectively). None of the subjects in both the groups who consumed alcohol+betelnut or smoking+betelnut were HPV positive. Even though 16.6% of the subjects in the OSCC group had no history of exposure to smoking, chewing betelnut or consumption of alcohol, 40% of such subjects were HPV positive [Table/Fig-4].
Relation of HPV positivity to risk-factors such as betelnut chewing, smoking and alcohol consumption among high risk and OSCC groups.
| Habits (%) | Groups |
|---|
| High risk (%) | OSCC (%) |
|---|
| Smoking | No | 80 | 86.7 |
| Yes | 20 | 13.3 |
| HPV +ve | 33.3 | 25 |
| Alcohol | No | 90 | 100 |
| Yes | 10 | 0 |
| HPV +ve | 66.6 | 0 |
| Betelnut | No | 83.4 | 76.7 |
| Yes | 16.6 | 23.3 |
| HPV +ve | 20 | 28.5 |
| Smoking + Alcohol | No | 56.7 | 63.4 |
| Yes | 43.3 | 36.6 |
| HPV +ve | 61.5 | 63.6 |
| Smoking + Betelnut | No | 100 | 100 |
| Yes | 0 | 0 |
| HPV +ve | 0 | 0 |
| Alcohol + Betelnut | No | 90 | 95.4 |
| Yes | 10 | 6.6 |
| HPV +ve | 0 | 0 |
| Smoking + Alcohol + Betelnut | No | 100 | 96.7 |
| Yes | 0 | 3.3 |
| HPV +ve | 0 | 100 |
| None | Yes | 0 | 16.6 |
| HPV +ve | 0 | 40 |
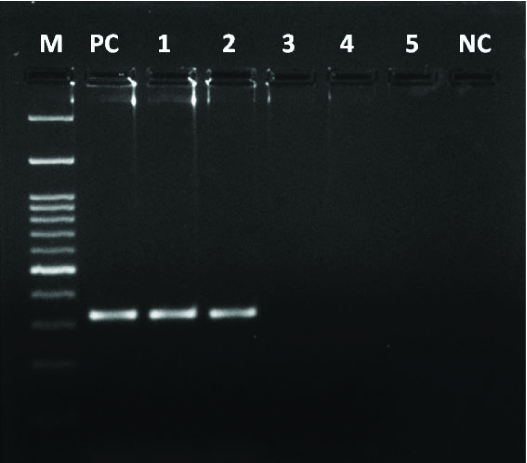
Genotyping was done as a part of the study to identify the high-risk subtypes of HPV. HPV subtype 16 was most commonly found in the study population (23/90) and was detected as an amplified product of 467bp [Table/Fig-5]. Similarly, HPV subtype 18 was also found in few samples (6/90) which were detected as an amplified product of 322bp [Table/Fig-6].
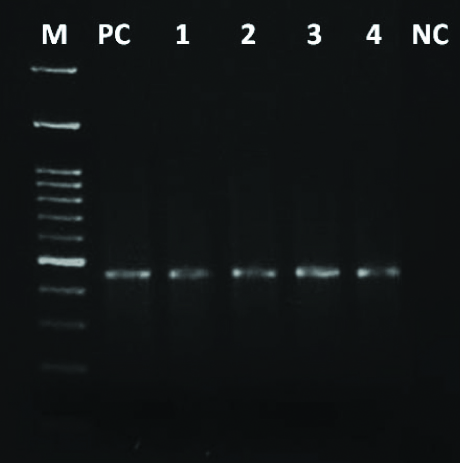
Genotyping of HPV 16 using primers amplifying a 467bp region. (M= 100bp marker, PC= Positive control, Lane 1-4= Positive samples, NC= Negative control).
Genotyping of HPV 18 using primers amplifying a 322bp region (M= 100bp marker, PC= Positive control, Lane 1-2= Positive samples, Lane 3-5= Negative samples, NC= Negative control).
Discussion
The present was study aimed at detecting and genotyping HPV in the saliva samples of OSCC, high risk and the normal healthy subjects by nested PCR. The results showed that subjects in the high risk group had similar HPV positivity numbers when compared with OSCC subjects. Furthermore the results indicated that subjects who smoked and consumed alcohol had 61.5% and 63.6% of HPV positivity respectively. About 16.6% of OSCC subjects did not have any habits categorised as risk-factors but still 40% of them were HPV positive. HPV infection was present in 46.6% of the high risk subject’s and13.3% of the normal healthy subjects who were not exposed to any risk-factors which shows that even healthy subjects are prone to develop OSCC.
A similar pattern of observations were reported by Modak H et al., where 47% of the study population were HPV positive [17]. Another study by Kulkarni SS et al., showed that 70.6% of the OSCC subjects were HPV positive and the prevalence of HPV-18 was slightly higher than HPV-16 [18]. In the present study HPV-16 was more common than HPV-18 (92.3% vs 7.6% and 71.4% vs 28.5% in OSCC group and high risk group respectively). In contrast, a hospital based study by Laprise C et al., involving 350 subjects did not even find a trace of HPV in any of the samples [19]. This was quite in contrast to other similar studies including ours which used the same L1 specific PGMY09/PGMY11 primers for the detection of HPV.
In the countries worldwide, there has been a remarkable increase in the incidence of oropharyngeal cancers attributed to HPV [10,20,21]. All these data suggest that, in a country like India with diverse cultural and geographical differences, there is still need for standard sample collection and analytical methods for HPV detection. Our study genotyped only HPV-16 and 18, whereas other common subtypes and multiple infections might also be involved in the HPV infection which were not taken into consideration. There is not enough evidence suggesting that HPV could be transmitted through close contact such as kissing and shared eating, but it is still not out of question as significant number of studies have demonstrated the presence of high risk HPV in the saliva.
Saliva has been very promising as a screening tool for the diagnosis of various disease conditions in the last decade [22]. Utilisation of saliva has already benefited the medical field as it eliminates the painful invasive procedures such as repeated blood draws and biopsies. Since the salivary diagnostics is non-invasive, it allows the patients to collect their own samples and has a positive impact on patient compliance [23]. Several studies have demonstrated the utility of saliva in the molecular diagnosis of cancer. A study by Wasserman JK et al., showed that saliva of 47% of the study population was tested positive for HPV DNA. Along with HPV positivity, they also showed that it is specific for p16 positive tumours [24]. Another study by Wang Y et al., used saliva to detect HPV infection and somatic mutations in patients with Head & Neck cancer and concluded that DNA in saliva is a valuable biomarker [25]. Saliva could be a possible screening tool for testing HPV but many studies have questioned its sensitivity and specificity. Nevertheless, utilising saliva is a boon to the medical diagnostics and further research is required to define more possibilities for the utilisation of the same.
Limitation
Even though our study evaluated the feasibility of nested PCR to detect HPV infection in the saliva successfully, it still had some limitations. Firstly, the OSCC subjects were not categorized based on the site and stage of the malignancy. Secondly the detection of HPV DNA in saliva was considered as positive for HPV infection whereas inclusion of in-situ hybridization or immunohistochemistry with p16 antibody as a comparative assessment would have supported the specificity of the assay.
Conclusion
Our study utilised saliva for the detection of HPV using a nested PCR and validated that saliva could serve as a non-invasive tool in screening of HPV associated oropharyngeal cancers. This could replace the use of invasive biopsies for detection of the virus in the cancer subjects. The results suggested that subjects who were exposed to risk factors such as smoking, chewing betelnut and consuming alcohol are equally prone to HPV infection and development of OSCC.