Introduction
Oral Potentially Malignant Disorder (OPMD) is considered as a disease symptom which if neglected without treatment may lead to cancer. OPMDs comprise leukoplakia, erythroplakia, erytholeukoplakia (the combination of both leukoplakia and erythroplakia), Oral Submucous Fibrosis (OSMF) and lichen planus (which has a low risk of turning malignancy). It is observed that a variety of factors influence the spread of OPMDs and these includes age distribution, gender distribution and geographic distribution, amongst others.
Aim
The aim of the study is to identify the differential presence of p16 gene promoter hypermethylation in the saliva of smokers and non smokers.
Materials and Methods
A total of 52 saliva samples were collected and prepared with full acknowledgement of the subjects. DNA isolation, restriction digestion of genomic DNA, extraction of restriction enzyme digested genomic DNA, Polymerase Chain Reaction (PCR) and finally Agarose Gel Electrophoresis (AGE) were performed.
Results
14 (70%) out of 20 samples collected from smokers were found to be methylated in p16 gene while 6 (30%) out of 20 showed no methylation. In non smokers, 29 (91%) out of 32 samples were found to be methylated. The present study shows a marginally lower rate of p16 gene hypermethylation in smokers compared to non smokers.
Conclusion
p16 gene hyper methylation is not recommended to be used as a marker for early detection of OPMDs among Malaysians. Promoter hypermethylation of p16 gene is not an early event in the prognosis of Malaysian Oral Squamous Cell Carcinoma (OSCC) patients.
Introduction
An OPMDs is described as a disease symptom which if neglected without treatment may result in cancer. It occurs in about 2.5% of the general population and is an important aspect connected with the efforts of cancer prevention [1]. It is also described as an alteration of tissues structure in which neoplasm may occur. It is known that in many cases OPMDs do not become cancer. These OPMDs mostly occur in the buccal mucosa and in oral commissures [2]. The types of premalignancy which occur normally are leukoplakia, erythroplakia, erytholeukoplakia (the combination of both leukoplakia and erythroplakia), OSMF and oral lichen planus (which have a low risk of turning malignant). They are classified mostly depending on their clinical appearances or through histopathological diagnosis coupled with local factors. The most common premalignancy is Leukoplakia [1,3]. Various risk factors are listed in oral cancers, however, the most important ones are age distribution, gender distribution and geographic distribution. However, age is not considered as a major risk factor for OPMDs since, it occurs mostly in middle aged people (37-59 years). Incidence of such phenomena is around five percent in population below 30 years of age [1]. Oral cancer is rated as the fifth most common cancer in men and seventh most common cancer in women. In most countries, oral cancer is more prevalent in men than in women [4].
High risk countries are Bangladesh, Brazil, Cuba, Hungary, India, Melanesia, Pakistan, Papua New Guinea, Puerto Rico, Slovakia, Slovenia, Sri Lanka and Uruguay. United Kingdom (UK) is reported to have low incidence of oral cancers [4].
Oral squamous cell carcinoma: OSCC is a white or red or white and red mixed lump, seen on the lips or part of the tongue which lasts for more than three weeks. OSCC develops from premalignant lesions such as leukoplakia and oral squamous fibrosis. OSCC is the most frequent cancer of Head and Neck Squamous Cell Carcinoma (HNSCC). Worldwide, it is the sixth most common cancer. There are 7,80,000 new cases of HNSCC reported worldwide each year and over 3,00,000 or more die annually [5]. Among the new cases of oral cancers diagnosed, males form 68%. Deaths due to oral cancers occurred at a male to female ratio of 7:3 [6].
Etiology of oral squamous cell carcinoma: The risk factors of OSCC are partially associated with the genetic make up of individuals. In young age, the risk factors are low. The risk factors include the use of tobacco, and consumption of alcohol. Alcohol has no direct carcinogenic effect but acts in combination with other factors [7]. OSCC occurs mostly in elderly people from the age of 50. Certain chronic inflammations such as irritable bowel syndrome, atrophic gastritis may also lead to OSCC [8].
Diagnosis of oral squamous cell carcinoma: OPMDs detection at early stages helps in decreased morbidity of patients suffering from OSCC. It can be diagnosed through tools such as brush biopsy, endoscopy and oesophagoscopy which are employed to detect primary cancer. Chest X-ray and CT scan are in rich use on the detection of oral cancer. Deoxyribonucleic Acid (DNA) image cytometry is used to differentiate the normal epithelial cells from the malignant oral mucosal cells [3,9].
Epigenetics and methylation: DNA methylation is an epigenetic modification process having a methyl group (CH3) at the fifth carbon position of the pyrimidine ring of cytosine at Cytosine-Guanine (CpG) dinucleotide. Methylation takes place when the gene behaviour is altered. Methyl group is added to the CpG region in the presence of DNA Methyltransferase (DNMT) aided by co enzyme S-adenosyl methionine to 5“C of a CpG dinucleotide which automatically get methylated. It alters the binding of histone complex and prevents DNA transcription [10]. Aberrant DNA methylation at the CpG site occurs so as to cause genome instability resulting in the development of cancer [11].
CpGs are not randomly distributed throughout the genome but are oriented at CpG enriched regions [12] known as CpG islands. It has a sequence greater than 0.5 kb with a G+C content higher than or equal to 55%. They are 0.5-5 kb in length and occur one in every 100 kb of the genome. These CpG islands are often disproportionately oriented in the 5“ promoter regions of genes. Around 50% of all human genes have their CpG islands in the promoter region. Promoter associated CpG islands are generally not methylated; although, methylation of subgroups of CpG islands often occurs [12].
Cancer cells frequently exhibit localised methylation of promoter CpG islands. Hypermethylation is generally known by the presence of a methyl group on the promoter region of a tumour suppressor gene. It leads to the reduction of tumour suppressor gene associated with several types of cancers [13]. The promoter region of hypermethylation is associated with the inactivation of DNA transcriptional silence together with the loss of expression of tumour suppressor. Approximately 100-400 hypermethylated CpG islands are present in the promoter regions of most tumours [12]. Genes involved in cell cycle control, DNA repair, carcinogen metabolism, cell to cell interactions, apoptosis and angiogenesis are actively interfered by hypermethylation [12]. It interacts differently in different genes and influences the genetic lesions. It often develops in the early stages of tissue preceding cancer, and then progressively increases during carcinogenesis. Some CpG islands are prone to large scale epigenetic deregulation. The tumour suppressor genes in OSCC are known to help the cell cycle and assist the cell growth. It keeps the cells from dividing too fast or in an uncontrollable way during DNA transcription [14]. The hypermethylation tumour suppressor genes present in OSCC are p16 (p16INK4a), O6-methylguanine-DNA-methyltransferase (MGMT), Death Associated Protein-Kinase (DAP-K) and TSG p53 CDH1 (Cadherin 1 gene).
In the present work, it is attempted to compare the hypermethylation of tumour suppressor genes in the saliva of smokers and non smokers of chosen samples from Malaysia. It is also aimed at identifying hypermethylation markers in OPMDs for early detection of OSCC.
Methodology
Preparation of saliva samples: This work was cleared by the ethical committee of Northumbria University, UK and MAHSA University, KL, Malaysia. All the necessary and required ethical procedures were followed during the sample collection and analysis. Informed consent was obtained in writing from all the participating subjects after explaining them the aims and objectives of the study in detail. All the samples collected were given a barcode and their individual information was kept confidential. Samples of saliva containing buccal epithelial cells were collected from 52 subjects from three different universities in Malaysia. The source of these samples were 3 (5.8%) from City University, 7 (13.5%) from UCSI University and 42 (80.8%) from MAHSA University. Samples were collected during the period between 13 February 2015 to 2 March 2015. The profiles of the samples include 23 female non smokers, nine male non smokers, one female smoker and 19 male smokers. The dearth in the female smoker cohort is in account of the strict cultural and ethical considerations prevalent in the Malaysian society. Approximately 1 mL of saliva was collected from each individual. They were asked to rinse their mouth five times with water (to remove food debris) and to scrap their buccal mucosa with a new set of toothbrush so, as to get a quality saliva sample containing buccal epithelial cells. The saliva samples were collected in sterile containers. The samples collected outside the laboratory were placed in a sealed plastic bag and transported in dry ice to the laboratory.
DNA extraction: The DNA from the saliva collected was extracted from 500 μL sample. This was added with 50 μL detergent solution to destabilise and break the membrane of the cells and release the contents. To this, 200 μL protease enzyme was added to remove the protein. It was then repeatedly washed with 200 μL of chloroform to remove the fat bodies. Three drops of 2M salt solution was then added to precipitate and solidify the DNA. The DNA was then precipitated by adding few drops of absolute ethanol. Since, the DNA is hydrophilic, it was resuspended in 200 μL distilled water. A known quantity (10 μL) of the resultant suspension was taken in a 1.5 mL eppendorf tube containing 900 μL distilled water. The quantity of the DNA was analysed calorimetrically using spectrophotometer. The purity of the DNA was evaluated based on the ratio of OD260/280. Quantification of all DNA samples was made using a SECOMAM UViline 9400 spectrophotometer at 260 and 280 nm absorbance.
Restriction digestion of genomic DNA: Restriction digestion of genomic DNA using restriction enzymes Hpa II was carried out using a kit of Promega Corporation. Restriction enzyme was stored at -20°C to preserve the activity by following the protocols of the manufacturer. The purified genomic DNA was digested with restriction enzymes Hpa II. The reaction mix include 2 μL of 10 x buffer (pH-7.4), 0.5 μL BSA, 0.5 μL restriction enzyme Hpa II, 10 μL purified DNA and 7.0 μL distilled water making up to a total volume of 20 μL for each tube. The samples were incubated in a water bath maintained at 37°C for one hour. The DNA was further purified by chloroform wash using 100 μL of chloroform and then centrifuged at 13000 rpm for five minutes followed by ethanol precipitation using 50 μL of absolute alcohol with 0.5 μL of 2M salt solution. It was again centrifuged. The supernatant was removed and the pellet in the tube is dried and suspended in 10 μL of distilled water for further testing.
Polymerase chain reaction: Primers were designed to recognise CpG rich promoter region (510 bp upstream) of transcription start site enclosing Hpa II restriction site. The internal control was the nested primer which is annealed outside the restriction site so that its product remains unaffected by the restriction digestion using Hpa II. The semi nested multiplex PCR was carried out in a 50 μL reaction mixture consisting 25 μL of 2 x PCR master mix, primers stock (p16S, p16A and p16N each 5 μL), 5 μL DNA template, 5 μL distilled water, 3mM MgCl2, 0.06U of Taq and 400 μM of each dNTPs (1st BASE Pvt. Ltd). The sequences of the PCR primers are given in [Table/Fig-1].
Primer sequence for p16 gene synthesised (Medox).
| Oligo | Sequence 5′-3′ | Tm C | Mer | GC (%) | OD | Molecular weight (G/mol) | Molar mass (nM) | Mass amount (gm) | Volume for 100 pmol |
|---|
| P16S | GAAGA-AAGAG-GAGG-GGCTG | 58.8 | 19 | 57.9 | 9.5 | 6015 | 39.3 | 237 | 393 |
| P16A | GGTCG-GGTA-GAGGA-GGTGC | 63.1 | 19 | 68.4 | 9.1 | 5989 | 41.8 | 251 | 418 |
| P16N | GCGCT-ACCTG-ATTCC-AATTC | 57.3 | 20 | 50 | 11.1 | 6027 | 54.3 | 327 | 543 |
The PCR program consisted of 30 amplification cycles, each consisting of initial denaturation at 56°C for 30 sec, denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec, final extension at 72°C for five minutes and soak at 40°C for 57 min. This is a modified PCR method [15]. The amplified product was separated using two percent agarose for 35 min at 50 V. Two bands indicate the presence of methylation and a single band indicate the absence of methylation.
Results
The number of subjects and other associated details (age, gender and smoking habit) of the people from whom the samples were collected are given in [Table/Fig-2]. A total of 52 subjects were used in this study. The number of males 28 (53.8%) sampled in this study were marginally higher than the females 24 (46.2%). All the subjects were in the age group of 19-48 years. The fragment size of the methylated and unmethylated amplicons observed in p16 gene was 510 bp and 340 bp where the hypermethylation took place. The internal control was 170 bp [Table/Fig-3]. A view of the agarose gel obtained from the AGE analysis on the methylation and non methylation of the DNA from the samples collected from smokers and non smokers is given in [Table/Fig-3]. The amplicons were electrophoresed in 2% agarose gel and stained with ethidium bromide.
Methylated and unmethylated genes according to gender and smoking characteristics.
| Cases | Samples (n) | Methylated p16, n (%) | Unmethylated p16, n (16%) |
|---|
| Gender |
| Female | 24 | 23 (96%) | 1 (4%) |
| Male | 28 | 20 (71.4%) | 8 (28.6%) |
| Total | 52 | 45 (87%) | 7 (13%) |
| Smoking status |
| Smokers | 20 | 14 (70%) | 6 (30%) |
| Non smokers | 32 | 29 (91%) | 3 (9%) |
Gel doc image indicating the band intensity of promoter methylation analysis of p16 gene from the sample obtained.
U: Unmethylated; M: Methylated. The arrow negative to positive shows the electrophoretic motion. The numbers indicate the lanes. The double bands indicate methylation.
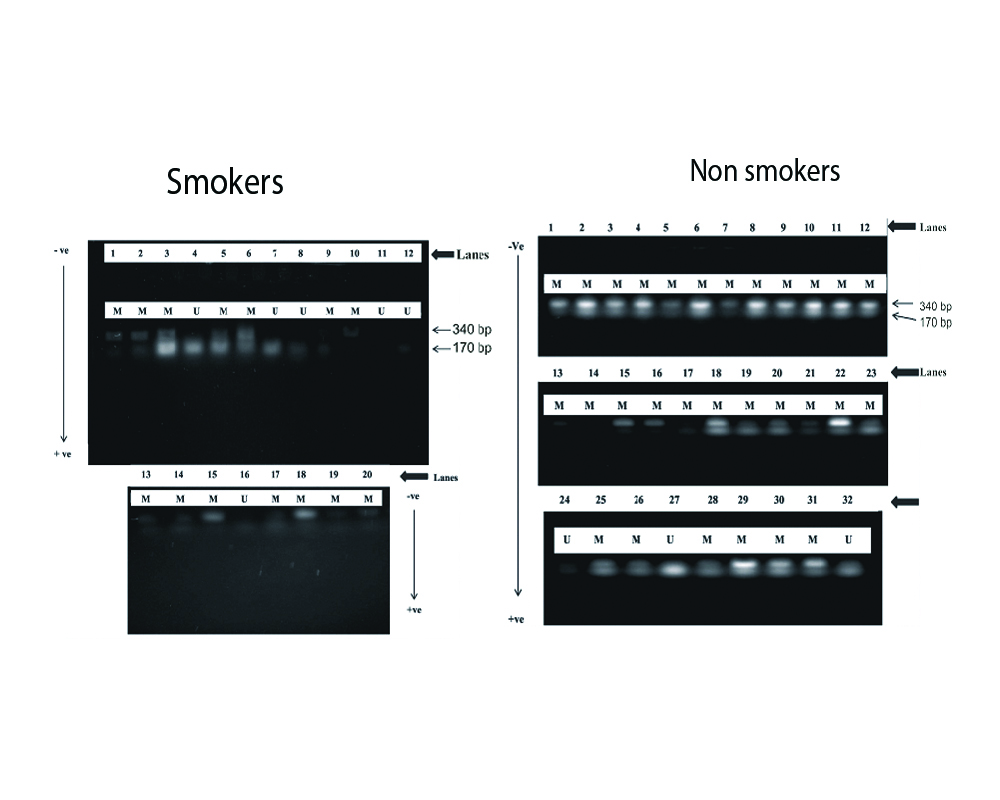
The methylation status of p16 gene of smokers and non smokers were analysed and recorded. The p16 gene was observed to be methylated in 45 (87%) samples and unmethylated in 7 (13%) samples irrespective of being smokers or non smokers. The smokers showed 14 (70%) hypermethylation of p16 gene when compared to 29 (91%) of the non smokers. The study also showed that 23 (96%) of the samples from females were methylated when compared to 20 (71.4%) of the males. The percentage occurrence of methylated and unmethylated samples with reference to gender and smoking status are given in [Table/Fig-2].
Discussion
Previous studies on p16 promoter region hypermethylation of oral rinse saliva samples collected from smokers were more conspicuous than that of non smokers [16,17]. There appears to have a relationship between the smoking habit and p16 gene promoter hypermethylation; however, it is not consistent in the sense that p16 gene cannot be used as an early indication in the genesis of OPMD among Malaysians. The results obtained in the present study indicate that the incidence of methylation of p16 gene was relatively low (70%) among smokers whereas in non smokers the methylation of p16 gene was high (91%). This may be accounted by the fact that Malaysian cuisine uses garlic in huge amounts. Any Malaysian meal is not complete without intake of garlic in liberal quantity. The chemical compound allicin present in garlic (Allium sativum) is a proven anti carcinogenic agent. Previous studies have shown that gene methylation pattern in mucous collected from smokers and non smokers were quite similar since the methylation of p16 gene was not uniform and they also showed that p16 gene occasionally got methylated in normal mucosa during aging [18].
Normal decision on any biopsy is based on the emergence of clinical risk factors which include site, size and the appearance of the lesions, smoking habit and OSCC history of the person. The criteria clinically adopted are far from adequate for the high risk OPMDs identification because of two major problems i.e., OPMDs could be easily mistaken for reactive or inflammatory lesions of non malignant potential (this means that no biopsy for these lesions is needed in most of the cases) and too many OPMDs on early diagnosis of cancer are not feasible [19].
Exposure to cigarette smoke and DNA methylation: Cigarette smoking is often considered to be one of the most robust environmental factors of DNA methylation. Though, it is well publicised that smoking is harmful to health, the practice is continuing unabated [20]. DNMT1 content has been modulated both at the transcription and at protein level. The enzymatic activity differs in different cell types. The reports showed that carcinogens present in cigarette such as arsenic, chromium, formaldehyde, polycyclic aromatic hydrocarbons and nitrosamines may lead to the damage of DNA by breaking its double-strands, as shown in the mouse embryonic stem cells which were directly exposed to cigarette smoke [21,22]. Other studies showed that smoking altered DNA methylation indirectly through DNA binding proteins such as Sp1 which were activated by cigarette smoke [23]. Sp1 is a transcription factor that commonly binds to GC-rich motifs in gene promoters and plays a key role in their early development. This appears to prevent a de novo methylation of CpGs [24]. Such may be the case in the present study as methylation was more prominent and had a higher percentage in non smokers (91%) than the smokers (70%). Furthermore, cigarette smoking may alter DNA methylation through hypoxia, as cigarette smoke contains carbon monoxide which binds to haemoglobin and compete with oxygen leading to a decrease in tissue oxygenation. Thus, hypoxia leads to the upregulation of dependent HIF-1 of methionine adenosyltransferase 2A an enzyme that synthesises S-adenosylmethionine, a biological methyl donor which is largely responsible for DNA methylation processes [17,20].
The inferences on DNA methylation are the regulation of gene expression and for the preservation of genomic integrity [25]. DNA methylation and modification of histones is most often considered as a mechanism involved in the regulation of epigenetic memory in mammalian cells. Along with the genetic disorders, epigenetic abnormalities play an active role in gene deregulation in cancer [26].
P16 promoter hypermethylation and oral cancer: Smoking habit changes the DNA methylation of p16 promoters, thereby causing changes in epigenetic expression of p16 gene comparing it with that of the non smokers and their percentage. The alterations in p16 gene are known to impact the cell cycle regulation, especially when the G1 phase is suppressed [27]. Several studies demonstrate the effect of p16 methylation in premalignant oral lesions with or without epithelial dysplasia and its association with advanced OSCC [28]. No association was observed on the p16 gene alterations with reference to the stages of cancer. Screening of both cyclin D1 and p16 aberrations in OSCCs may be useful in identifying aggressive tumours and disease recurrence in patients with a low prognosis. This shows that the expression of p16INK4a is associated with OSCCs [29]. The results obtained by present study, are in agreement with findings of a contemporary study in saliva samples [30].
Limitation
The study needs to be carried out on a larger sample scale to know more about the abnormalities of p16 gene hypermethylation in Malaysian populations. In the present study, the sample region is limited to the KL, region of Malaysia. Further studies need to be carried out across different age and ethnic cohorts to get a reliable estimate of the extent of hypermethylation. The near absence of female smokers because of the conservative social, cultural and religious practices is an important limitation even though it is not the primary focus of present study.
Conclusion
The result of this research showed a lower rate of p16 gene hypermethylation in smokers. It is possible that the p16 gene may not play a very important role in the genesis of OPMDs in Malaysian populations. Therefore, p16 gene hypermethylation cannot be used as a marker for early detection of OPMDs in Malaysia. The study also demonstrates that it is possible to isolate pure DNA through a localised medium from saliva.
Further work may be required to enhance the understating of the role of p16 gene in potentially malignant and malignant oral lesions. High risk patients may then be monitored to ensure the condition which does not progress to an oral cancer. Help of vaccines may also be resorted in future for OPMD patients.
[1]. Cornelio S, Rodrigues GS, A brief review of common oral premalignant lesions with emphasis on their management and cancer preventionSpringer 2011 73(4):256-61.10.1007/s12262-011-0286-622851837 [Google Scholar] [CrossRef] [PubMed]
[2]. Mahawar P, Anand S, Sinha U, Screening for pre-malignant conditions in the oral cavity of chronic tobacco chewersNat J Comm Med 2011 2(1):82-5. [Google Scholar]
[3]. Messadi DV, Diagnostic aids for detection of oral precancerous conditionsInt J Oral Sci 2013 5(2):59-65.10.1038/ijos.2013.2423743617 [Google Scholar] [CrossRef] [PubMed]
[4]. Warnakulasuriya S, Living with oral cancer: Epidemiology with particular reference to prevalence and life -style changes that influence survivalOral Oncol 2010 46(6):407-10.10.1016/j.oraloncology.2010.02.01520403722 [Google Scholar] [CrossRef] [PubMed]
[5]. Ovchinnikov DA, Cooper MA, Pandit P, Coman WB, Cooper JJ, Keith P, Tumour-suppressor gene promoter hypermethylation in saliva of head and neck cancer patientsTrans Oncol 2012 5(5):321-6.10.1593/tlo.1223223066440 [Google Scholar] [CrossRef] [PubMed]
[6]. Jensen A, Oral squamous cell carcinoma: An atypical presentation mimicking temporomandibular joint disorderJ Can Chiro Asso 2004 48(4):266-72. [Google Scholar]
[7]. Muange P, Chindia M, Njiru W, Dimba E, Mutave R, Oral squamous cell carcinome: A 6-month clinico-histopathologic audit in a Kenyan populationOpen J Stomat 2014 4:475-83.10.4236/ojst.2014.410064 [Google Scholar] [CrossRef]
[8]. Bodner L, Manor E, Friger MD, Waal VD, Oral squamous cell carcinoma in patients twenty years of age or younger-review and analysis of 186 reported casesOral Oncol 2014 50(2):84-89.10.1016/j.oraloncology.2013.11.00124296165 [Google Scholar] [CrossRef] [PubMed]
[9]. Yang WCV, Chung HR, Wu JY, Chen Yi, Wang DJ, Lee SY, Potential biomarkers for the cytologic diagnosis of oral squamous cell carcinomaJ Dental Sci 2010 5(2):60-9.10.1016/S1991-7902(10)60010-4 [Google Scholar] [CrossRef]
[10]. Miyamoto K, Ushijima T, Diagnostic and therapeutic applications of epigeneticsJap J Clin Oncol 2005 35(6):293-301.10.1093/jjco/hyi08815930038 [Google Scholar] [CrossRef] [PubMed]
[11]. Koo KM, Wee EJH, Rauf S, Microdevices for detecting locus-specific DnNA methylation at CpG ResolutionSci direct 2014 56:278-85.10.1016/j.bios.2014.01.02924514080 [Google Scholar] [CrossRef] [PubMed]
[12]. Esteller M, Epigenetics in CancerN Eng J Med 2008 358:1148-59.10.1056/NEJMra07206718337604 [Google Scholar] [CrossRef] [PubMed]
[13]. SupiC G, Kozomara R, Brankovic-Magic M, Jovic N, Magic Z, Gene hypermethylation in tumour tissue of advanced oral squamous cell carcinoma patientsOral Oncol 2009 45(12):1051-57.10.1016/j.oraloncology.2009.07.00719665921 [Google Scholar] [CrossRef] [PubMed]
[14]. Mehrotra R, Gupta A, Singh M, Ibrahim R, Application of cytology and molecular biology in diagnosing premalignant or malignant oral lesionsMol. Can 2006 5:1110.1186/1476-4598-11-5722905981 [Google Scholar] [CrossRef] [PubMed]
[15]. Solomon PR, Munirajan AK, Tsuchida N, Muthukumarasamy K, Rathinavel A, Selvam GS, Promoter hypermethylation analysis in myelodysplastic syndromes: Diagnostic and prognostic implicationInd J Med Res 2008 127:52-7. [Google Scholar]
[16]. Kulkarni V, Saranath D, Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissuesOral Oncol 2004 40(2):145-53.10.1016/S1368-8375(03)00143-X [Google Scholar] [CrossRef]
[17]. Liu M, Feng L, Tang X, Guo S, Gene promoter hypermethylation In Leukoplakia of the oral mucosaPathol Lab Med Int 2010 2:71-7.10.2147/PLMI.S10916 [Google Scholar] [CrossRef]
[18]. Roth MJ, Abnet CC, Hu N, Wang QH, Wei WQ, Green L, p16, MGMT, RAR±2, CLDN3, CRBP and MTIG gene methylation in esophageal squamous cell carcinoma and its precursor lesionsOncol. Rep 2006 15(6):1591-97.10.3892/or.15.6.159116685400 [Google Scholar] [CrossRef] [PubMed]
[19]. Li J, Molecular Assessment of Former Cancer Sites Predicts Second Oral Malignancy. Master’s in science, Honours [thesis]Lanzhou University 1988 [Google Scholar]
[20]. Lee KWK, Pausova Z, Cigarette smoking and DNA methylationFront Genet 2013 132(4):1-11.10.3389/fgene.2013.00132 [Google Scholar] [CrossRef]
[21]. Suter M, Abramovici A, Showalter L, Hu M, Shope CD, Varner M, In utero tobacco exposure epigenetically modifies placental CYP1A1 expressionMetabolism 2010 59(10):1481-90.10.1016/j.metabol.2010.01.01320462615 [Google Scholar] [CrossRef] [PubMed]
[22]. Huang J, Okuka M, Lu W, Tsibris JC, McLean MP, Keefe DL, Telomere shortening and DNA damage of embryonic stem cells induced by cigarette smokeRepr toxicol J 2012 35(1):89-95.10.1016/j.reprotox.2012.07.00322824788 [Google Scholar] [CrossRef] [PubMed]
[23]. Di YP, Zhao J, Harper R, Cigarette smoke induces MUC5AC protein expression through the activation of Sp1J Biol Chem 2012 287:27948-58.10.1074/jbc.M111.33437522700966 [Google Scholar] [CrossRef] [PubMed]
[24]. Han L, Lin IG, Hsieh CL, Protein binding protects sites on stable episomes and in the chromosome from de novo methylationMol Cell Biol 2001 21:3416-24.10.1128/MCB.21.10.3416-3424.200111313467 [Google Scholar] [CrossRef] [PubMed]
[25]. Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional up regulation of MAT2A in hepatoma cellsMol Cancer Therap 2011 10(6):1113-23.10.1158/1535-7163.MCT-10-101021460102 [Google Scholar] [CrossRef] [PubMed]
[26]. Jones PA, Baylin SB, The epigenomics of cancerCell 2011 128:683-92.10.1016/j.cell.2007.01.02917320506 [Google Scholar] [CrossRef] [PubMed]
[27]. Chien WW, Domenech C, Catallo R, Kaddar T, Magaud JP, Salles G, Cyclin-dependent Kinase 1 expression is inhibited by p16 (INK4a) at the the post-transcriptional level through the microRNA pathwayOncogene 2011 30(16):1880-91.10.1038/onc.2010.57021170085 [Google Scholar] [CrossRef] [PubMed]
[28]. Sailasree R, Abhilash A, Sathyan KM, Nalinakumari KR, Thomas S, Kannan S, Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinomaCancer Epidemiol Biomar Prev 2008 17(2):414-20.10.1158/1055-9965.EPI-07-028418268126 [Google Scholar] [CrossRef] [PubMed]
[29]. Uzawa N, Sonoda I, Myo K, Takahashi K, Miyamoto R, Amagasa T, Fluorescence in situ hybridization for detecting genomic alterations of cyclin D1 and p16 in oral squamous cell carcinomasCancer 2007 110:2230-39.10.1002/cncr.2303017893905 [Google Scholar] [CrossRef] [PubMed]
[30]. Kaliyaperumal S, Sankarapandian S, Evaluation of p16 hypermethylation in oral submucous fibrosis: A quantitative and comparative analysis in buccal cells and saliva using real-time methylation-specific polymerase chain reactionSouth Asian J Cancer 2016 5(2):73-9.10.4103/2278-330X.18164527275454 [Google Scholar] [CrossRef] [PubMed]