Materials and Methods
In this cross-sectional study, from February 2016 to March 2017, 97 S. aureus isolates obtained from various clinical infections from Sina hospital in Tabriz, Iran. Based on a previous study, and by considering α=0.05, β=0.2, power=80%, P1=0.61 and P2=0.32 and a difference of 10%, the sample size was estimated at 90 [8]. All detected S. aureus isolates confirmed by biochemical and genetic tests from patients referred to Sina hospital during study period included to the study. Exclusion criteria were species other than S. aureus and or duplicate isolates from the same patients. Of the total 97 S. aureus isolates, 87 were obtained from inpatients and 10 had out- patient source. The identity of all S. aureus isolates was confirmed by utilising the conventional bacteriological methods including Gram staining, catalase test, coagulase test, DNase test, mannitol salt agar growth, and 6.5% salt tolerance and later conventional Polymerase Chain Reaction (PCR) amplification was performed to verify species identification using the nuc gene as described previously [2,11]. The present study was approved by The Ethic Commission of Tabriz University of Medical Sciences (Number: 1394.930). Patients consent forms were obtained before sampling, forms were in Persian and all patients informed about procedure of sampling and study.
Antimicrobial Susceptibility Testing
Disc diffusion method was performed to determine antimicrobial susceptibility patterns of S. aureus in accordance with the recommendations of the Clinical and Laboratory Standards Institute (CLSI) 2016 guidelines [12]. Inoculum peparation was done in normal saline and all inoculums adjusted to 0.5 McFarland standards. Inocula were used in less than 15 minutes to prevent any changes in the number of bacteria. The antimicrobial agents tested were as follows: trimethoprim/sulfamethoxazole (25 μg), erythromycin (15 μg), cefazolin (30 μg), cefoxitin (30 μg), ciprofloxacin (5 μg), penicillin (10 μg), clindamycin (2 μg) and gentamycin (10 μg) (MAST Diagnostics, Merseyside, UK). Vancomycin susceptibility testing of S. aureus was performed by using vancomycin screen agar plates containing 6μg/mL vancomycin and vancomycin E-test according to CLSI 2017 guidelines [12,13]. MRSA isolates were detected by using oxacillin screening agar (plates had 4% NaCl and 6 mg/L of oxacillin) and cefoxitin disc diffusion test (30 ug) [14]. Staphylococcus aureus ATCC® 33591™, S. aureus ATCC® 25923™, S. aureus ATCC® 29213™, Enterococcus faecalis ATCC® 51299™, and E. faecalis ATCC® 29212™ were used as the control strains. For D test examination in isolates, erythromycin and clindamycin discs were placed adjacent to each other during antimicrobial susceptibility test. The growth of the S. aureus isolates up to the edges of the disc, flattening of the clindamycin zone near the erythromycin disc (resistant) was considered D test positive.
Biofilm Formation Assay with Microtiter Plate Method
S. aureus biofilm formation was analysed in 96 well flat bottom polystyrene plates (Greiner Bio One, Germany), under static conditions for 48 hours as previously described [15,16]. For biofilm development, inoculum of S. aureus equivalent to 107 Colony Forming Unit (CFU)/mL was prepared by adjusting culture grown bacterial suspensions in Trypticase Soy Broth (TSB) (Hi-media, India) from overnight cultures to an Optical Density at 600 nm (OD600) of 0.1 and further 100 μL of each adjusted inoculum was added to the wells. After 48 hours incubation at 37°C, plates were tenderly washed only once with 1x Phosphate Buffered Saline (PBS; pH 7.4) and stained with 100 μL of 0.1% Crystal Violet (CV) for 30 minutes at room temperature. Excess CV was expelled by washing, and CV stained biofilm was then solubilised in 200 μL of 95% ethanol and supernatant was transferred to a fresh microtiter plate. Biofilm was evaluated by measuring absorbance of the supernatant at 570 nm. Biofilm assays were performed in triplicate for each clinical strain and the mean biofilm absorbance quality was determined. OD of stained adherent bacteria were determined with a micro ELISA auto reader (model 680, Bio rad), and the wavelength of values was considered as an index of bacteria adhering to surface and forming biofilms. OD readings of wells with ethanol were used as blank and subtracted from all test values. Biofilm production was considered high, moderate, or weak as described previously [9].
DNA Extraction
DNA extraction was done by DNeasy kit (Qiagen Inc.) according to manufacturer’s instructions and boiling method [17]. The extracted DNA concentrations were determined by Nanodrop 1000 (NanoDrop, Wilmington, USA). One microliter of each DNA was used as template in the PCR reaction.
Detection of mecA Gene
DNA of S. aureus isolates with the concentration of 0.1 ng/μL was used as the templates for PCR analysis. Conventional PCR was carried out using CINNA GEN MASTERMIX (Cinnaclon, Tehran, Iran) and mecA primer as described previously [18]. The strain S. aureus ATCC® 43300™ (mecA positive) was used as positive control in this study. Amplification was carried out in an Eppendorf thermocycler (Eppendorf, Hamburg, Germany) as follows: initial denaturation at 94°C for five minutes, followed by 35 cycles of 30 seconds for denaturation at 94°C, 30 seconds for annealing at 55°C, and one minute for primer extension at 72°C, followed by terminal extension at 72°C for seven minutes [19]. Electrophoresis of PCR products was performed on 1% agarose gel using SYBR™ Safe DNA Gel Stain (Invitrogen) [20]. The stained gels were viewed on a UV transilluminator (Biorad, UK).
Detection of icaABCD Genes
To evaluate the biofilm formation, the presence of icaABCD genes was analysed by PCR amplification using specific primers as described previously [21]. PCR amplification was performed with an Eppendorf thermal cycler (Mastercycler® gradient). Amplification program consisted of initial denaturation at 94°C for five minutes, 30 cycles of denaturation at 94°C for 60 seconds, annealing at 55°C for 60 seconds (icaA), 52°C for 30 seconds (icaB), 55°C for 30 seconds (icaC), 55°C for 30 seconds (icaD) and extension at 72°C for 60 seconds with a final step of 72°C for 10 minutes [21]. The PCR products were analysed by electrophoresis in a 1.4% agarose gel using SYBR™ Safe DNA Gel Stain (Invitrogen).
Statistical Analysis
Statistical analysis was performed with SPSS program version 17.0 (SPSS, Chicago, IL, USA). The variables were analysed by univariate analysis using chi square or fisher’s exact test, as appropriate. Statistical significance was set at 0.05.
Results
In the present study, 99 isolate of S. aureus were collected from various clinical specimens comprising but for biofilm assay and genotyping, 97 S. aureus isolates were found suitable according to the study criteria. The mean age of the patients was 40.3±24 years and 55 (55.6%) patients were males. Source of the isolates were:blood (n=39, 40.2%), wound (n=50, 51.54% from skin, surgery, internal, burn and infectious wards), urine (n=5, 5.1%) and body fluids (n=3, 3.09%) from patients admitted to various wards including: infectious diseases (n=20, 20.61%), burn (n=18, 18.55%), intensive care unit (n=17, 17.52%), dermatology (n=14, 14.43%), internal (n=13, 13.4%), surgery (n=7, 7.21%) and out patients (n=8, 8.24%).
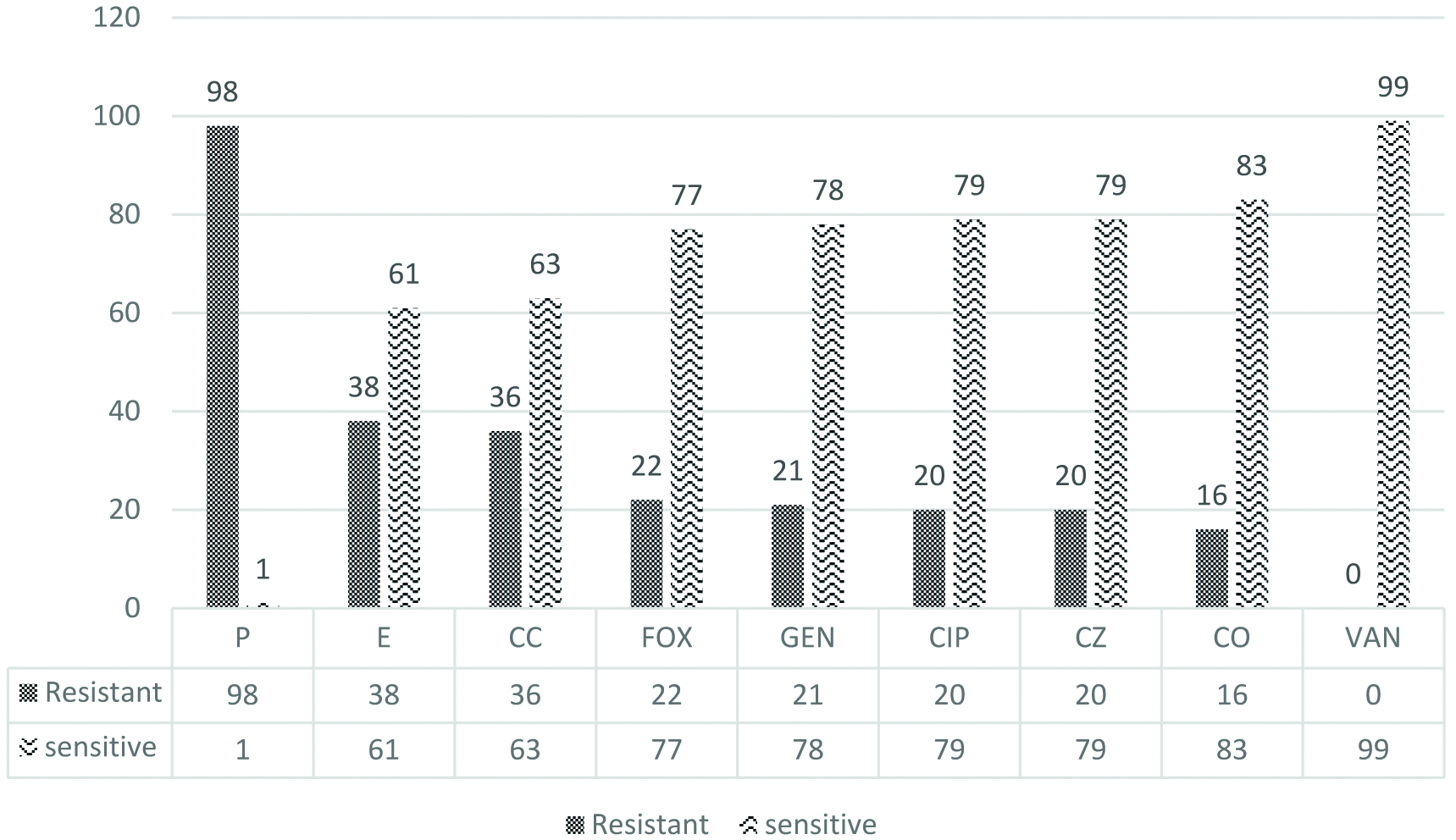
96 (98.9%) of the isolates were penicillin resistance followed by non susceptibility towards erythromycin (38.4%), clindamycin (36.24%), cefoxitin (22.2%), gentamycin (21.2%), ciprofloxacin, cefazolin (each 20.2%), and trimethoprim/sulfamethoxazole (16.2%). Of 36 clindamycin resistant isolates, 9 (25%) were D test positive. All S. aureus isolates were susceptible to vancomycin. 22 (22.68%) isolates were recognised phenotypically as Methicillin-Resistant Staphylococcus aureus (MRSA) by cefoxitin disk. Of these 22 isolates, 20 were obtained from inpatients and two from outpatients. The result was not significant at p>0.64. Of these MRSA isolates 18 (81.8%) of them possessed the mecA gene (p<0.001). AST patterns of S. aureus exhibits in [Table/Fig-1].
Antibiotic susceptibility pattern of Staphylococcus aureus isolates*. Y-axis represents percent of isolates, X-axis represents antibiotics tested.
P: Penicillin; E: Erythromycin; CC: Clindamycin; FOX: Cefoxitin; GEN: Gentamycin; CIP: Ciprofloxacin; CZ: Cefazolin; CO: Cotrimoxazole; VAN: Vancomycin
Of 97 S. aureus isolates, biofilm formation was studied in all isolates. Assessment of biofilm formation in these isolates presented five. 5 (15%) of the isolates as strong biofilm producer, while 28 (28.9%) displayed moderate biofilm formation, and 56.7% (n=55) showed weak biofilm formation. By phenotypic method nine isolates did not reveal biofilm production. Among 18 MRSA isolates confirmed by mecA gene [Table/Fig-2,3], 1 (9.09%) was strong producer, 4 (22.22%) were moderate producers and 12 (66.66%) of them were found to be weakly adherent while one isolate did not form biofilm. On the other hand, among MSSA isolates, 4 (5.1%) isolates were found to be strong producers, 24 (30.4%) were moderate and 43 (54.4%) were found weakly adherent. Overall, 88 (90.7%) of S. aureus isolates were biofilm producers [Table/Fig-4]. [Table/Fig-5] show the relation between mecA feature, biofilm capability and type of clinical specimen. Among various clinical sources, except two isolates from blood, all had shown ability to form biofilm. All isolates obtained from urine specimen had shown either weak (60%) or moderate (40%) biofilm producing ability. There was no relation between presence of icaABCD genes and biofilm formation (p=0.74).
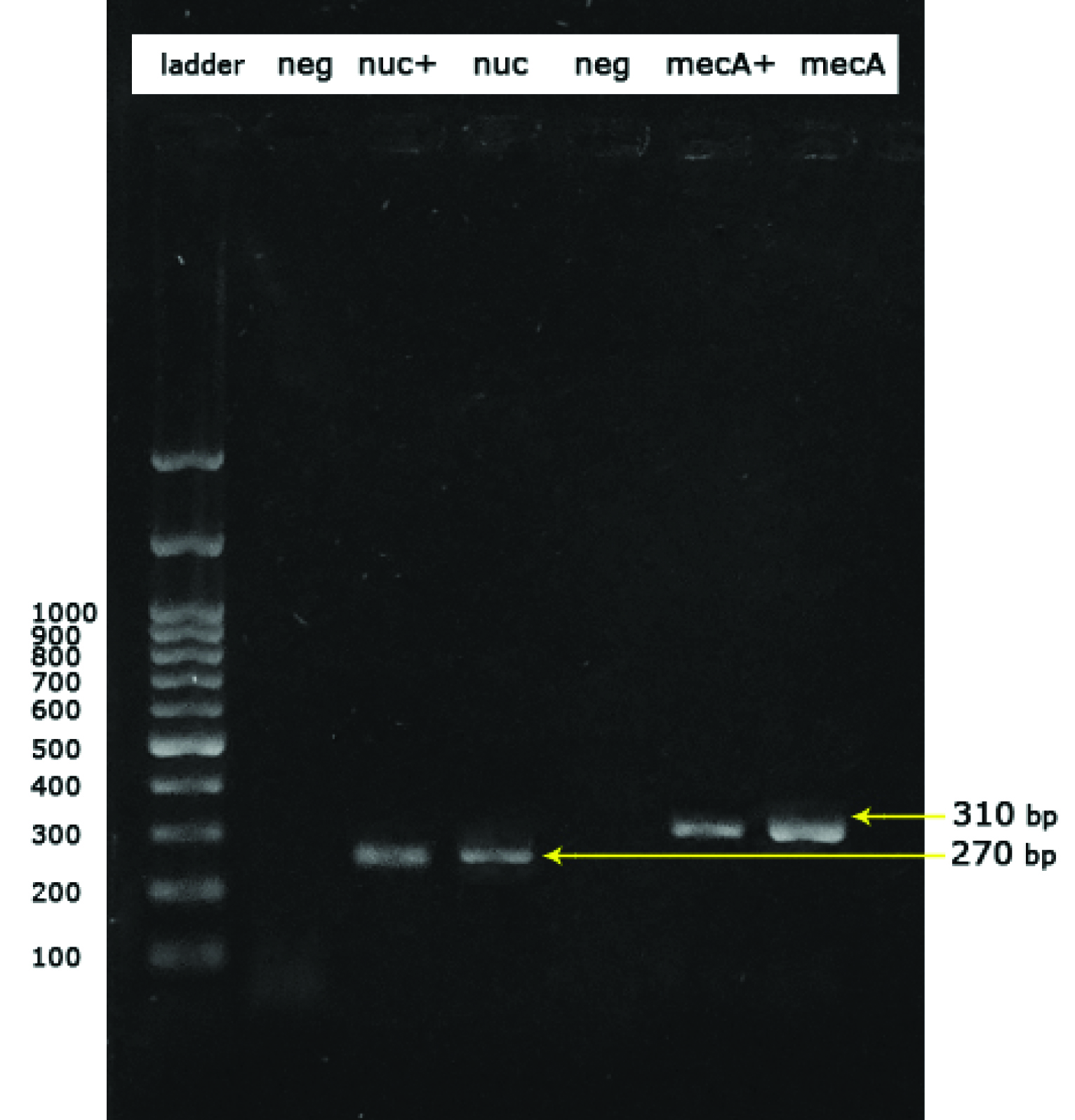
Screening S. aureus for the presence of nuc gene for confirmation of species and mecA gene for resistance to methicillin.
Lanes are respectively: ladder:Ladder 100bp, neg:Negative control (Escherichia coli ATCC® 25922 ™), nuc+:nuc positive control (S. aureus ATCC® 25923™), nuc:nuc positive sample (270 base pair), neg:negative sample, mecA+:mecA positive control (S. aureus ATCC® 43300™) (310 bp) and mecA:mecA positive sample (310 bp)
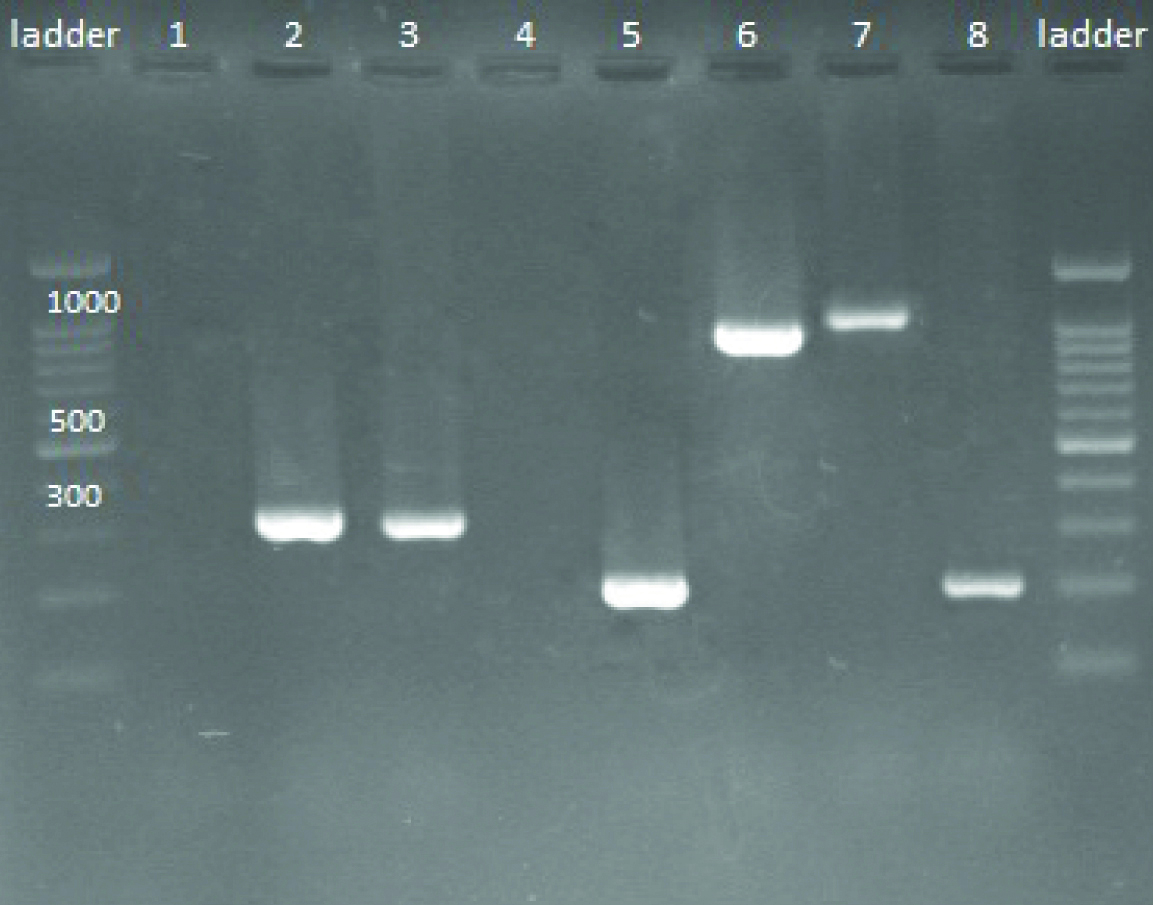
Screening of S. aureus isolates for the presence of icaABCD and mecA by PCR methods.
Lanes are respectively: ladder: Ladder 100 bp; 1: Negative control (Escherichia coli ATCC® 25922 ™); 2: mecA positive (310 bp (base pair); 3: mecA positive (310 bp); 4: Negative sample; 5: icaA positive (188 bp); 6: icaB positive (900 bp); 7: icaC positive (1100 bp); 8: icaD positive (198 bp)
Biofilm forming ability and biofilm genes involved in MRSA and MSSA isolates. p-value less than 0.05 was considered significant. Comparison groups were strong to moderate biofilm producers of MRSA and MSSA versus weak or negative ones.
| Genes | Biofilm producing ability in MRSAΨ | Total (%) | Biofilm producing ability in MSSAΨ* | Total (%) | p-value* |
|---|
| Strong | Moderate | Weak | Negative | Strong | Moderate | Weak | Negative |
|---|
| icaA | 1 | 4 | 12 | 1 | 18 (100) | 4 | 20 | 37 | 18 | 61 (77.2) | 0.827 |
| icaB | 1 | 4 | 8 | 0 | 13 (72.2) | 4 | 20 | 36 | 19 | 60 (75.9) | 0.561 |
| icaC | 1 | 2 | 6 | 9 | 9 (50) | 2 | 12 | 29 | 36 | 43 (54.4) | 0.915 |
| icaD | 1 | 4 | 13 | 0 | 18 (100) | 4 | 24 | 51 | 0 | 79 (100) | 0.535 |
ΨMRSA:Methicillin resistance S. aureus; MSSA:Methicillin sensitive S. aureus
*Descriptive analysis and chi square tests were applied for analysis
Cross tabulation between mecA characteristic, biofilm ability and type of clinical specimen.
| mecA | Clinical specimen | Biofilm ability | Total |
|---|
| Negative | Weak (+) | Moderate (++) | Strong (+++) |
|---|
| Positive | Blood | 0 | 5 | 2 | 1 | 8 |
| Wound (other than burns) | 0 | 2 | 1 | 0 | 3 |
| Burn wound | 1 | 5 | 1 | 0 | 7 |
| Total | 1 | 12 | 4 | 1 | 18 |
| Negative | Urine | 0 | 3 | 2 | 0 | 5 |
| Blood | 2 | 15 | 10 | 2 | 29 |
| Body fluids (other than blood and urine) | 0 | 2 | 1 | 0 | 3 |
| Wound (other than burns) | 6 | 17 | 8 | 2 | 33 |
| Burn wound | 0 | 6 | 3 | 0 | 9 |
| Total | 8 | 43 | 24 | 4 | 79 |
S. aureus isolates showing weak or moderate or strong biofilm formation were further analysed to possess biofilm genes and was 81 (83.5%), 71 (73.2%), 51 (52.5%), and 97 (100%) of them revealed icaA, icaB, icaC and icaD genes, respectively [Table/Fig-3]. All MRSA isolates were positive for icaA and icaD genes, while icaB gene was detected in 13 (72.2%) isolates and icaC being shown by 9 (50%) isolates. On the other hand, all MSSA isolates were positive for icaD gene only and 61, 60 and 43 isolates were observed positive for icaA, icaB and icaC genes respectively [Table/Fig-4,5]. However, there was no signifificant difference between MRSA and MSSA isolates for the presence of icaADBC operon (p=0.789).
Phenotypic and genotypic biofilm forming features of S. aureus isolates depicted in [Table/Fig-6] . All isolates irrespective of being MRSA or MSSA were positive for icaD gene. 9.09%, 6.84% and 5.77% of isolates were found negative for biofilm activity, nevertheless had icaA, icaB and icaC genes, respectively. On the other hand, 88.8%, 83.3% and 86.6% of S. aureus isolates were negative for icaA, icaB or icaC genes respectively, but had shown in vitro weak or moderate or strong biofilm activity.
Correlation between phenotype and genotype of biofilm production in clinical S.aureus isolates. Descriptive analysis and chi square tests were applied for analysis. p-value less than 0.05 was considered significant.
| Biofilm genes characteristics | Phenotypic biofilm ability | Total | p-value |
|---|
| Negative | Weak (+) | Moderate (++) | Strong (+++) |
|---|
| icaA | positive | 8 | 51 | 24 | 5 | 88 | 0.0053 |
| negative | 1 | 5 | 3 | 0 | 9 |
| Total | 9 | 56 | 27 | 5 | 95 |
| icaB | positive | 5 | 39 | 24 | 5 | 73 | 0.038 |
| negative | 4 | 16 | 4 | 0 | 24 |
| Total | 9 | 55 | 28 | 5 | 97 |
| icaC | positive | 3 | 32 | 14 | 3 | 52 | 0.766 |
| negative | 6 | 23 | 14 | 2 | 45 |
| Total | 9 | 55 | 28 | 5 | 97 |
| icaD | positive | 9 | 55 | 28 | 5 | 97 | NSΨ |
| negative | 0 | 0 | 0 | 0 | 0 |
| Total | 9 | 55 | 28 | 5 | 97 |
ΨNot significance (because of absence of negative icaD isolate)
When source of clinical specimen was compared with ability of isolate to form biofilm and presence of ica and mecA genes, interestingly no MRSA isolate obtained from urine and body fluids was positive either for icaB or icaC genes [Table/Fig-7].
A comprehensive view of icaB and icaC genes distribution among different clinical specimens and their correlation with mecA gene.
| mecA | ica genes | Clinical specimens | Total |
|---|
| Urine | Blood | Body fluids (other than blood and urine) | Wound | Burn wound |
|---|
| Positive | icaB | positive | 0 | 6 | 0 | 2 | 5 | 13 |
| negative | 0 | 2 | 0 | 1 | 2 | 5 |
| Total | 0 | 8 | 0 | 3 | 7 | 18 |
| Negative | icaB | positive | 4 | 22 | 3 | 26 | 6 | 61 |
| negative | 1 | 9 | 0 | 7 | 3 | 20 |
| Total | 5 | 31 | 3 | 33 | 9 | 81 |
| Positive | icaC | positive | 0 | 6 | 0 | 1 | 2 | 9 |
| negative | 0 | 2 | 0 | 2 | 5 | 9 |
| Total | 0 | 8 | 0 | 3 | 7 | 18 |
| Negative | icaC | positive | 3 | 20 | 1 | 14 | 6 | 44 |
| negative | 2 | 11 | 2 | 19 | 3 | 37 |
| Total | 5 | 31 | 3 | 33 | 9 | 81 |
Antibiotic susceptible and non susceptible S. aureus isolates had no significant difference in biofilm formation [Table/Fig-8].
Antimicrobial resistance pattern of S. aureus isolates and its relation with phenotypic biofilm features. Descriptive analysis and chi square tests were applied for analysis. p-value below 0.05 was considered significant.
| Antibiotics | Non susceptibility | Susceptibility | p-value |
|---|
| Biofilm former | Nonbiofilm former | Biofilm former | Non biofilm former |
|---|
| Erythromycin | 92.1% | 7.9% | 90.2% | 9.8% | 0.636 |
| Clindamycin | 91.7% | 8.3% | 90.5% | 9.5% | 0.859 |
| Cefazolin | 90% | 10% | 91.1% | 8.9% | 0.516 |
| Cotrimaxazole | 87.5% | 12.5% | 91.6% | 8.4% | 0.116 |
| Ciprofloxacin | 90% | 10% | 91.1% | 8.9% | 0.516 |
| Gentamycin | 90.5% | 9.5% | 91% | 9% | 0.64 |
| Vancomycin | 0% | 0% | 90.9% | 9.1% | NSΨ |
ΨNot significant (because of absence of vancomycin resistance isolate)
Discussion
S. aureus exploits many virulence factors, ability to adhere and form biofilm on host surfaces to attain the infectious level. The attachment and biofilm formation on abiotic surfaces like catheters and implanted devices are one of the most important virulence factors in S. aureus and is responsible for chronic or persistent infections [22]. In this regard, the phenotypic characterisation of adhesion and biofilm formation and related genetic elements involved in diverse clinical isolates of S. aureus might permit a better understanding of the complicated process of biofilm formation and infections caused by this microorganism [23]. Several studies have shown that formation of biofilm in S. aureus causing catheter associated and nosocomial infections is related to the presence of icaA and icaD genes [8,24]; however still lacunae exists, particularly in Iran for information regarding the source of bacterium compared with type of biofilm activity and the respective genes being involved. Moreover, research studies available have focused only on the presence of icaA and icaD genes or had restricted to only one source.
In the present study, all isolates were susceptible to vancomycin; resistance to vancomycin has been sporadically reported from some areas of the world, similar to Iran [25,26].
Microtiter plates were selected for biofilm formation assay and quantify attachment. Yet, presence and expression of biofilm genes ought to be confirmed by genotypic characterisation methods. Present study indicates a high prevalence of the icaADBC genes among S. aureus isolates. Since, biofilm protects microorganisms from opsonophagocytosis and antimicrobial agent as well as has a direct and indirect impact on healing process, through the production of destructive enzymes and toxin and promoting a chronic inflammatory state, presence of these genes provides vital information on the way of their pathogenesis [7].
In the present investigation study, 88 of the 97 S. aureus clinical isolates produced biofilm in vitro, and all the 88 isolates were found to possess the icaD gene. On the other hand, few isolates were observed to possess the ica genes but were negative on phenotypic test for biofilm formation and the vice versa condition was detected in the many isolates which furnished biofilm activity but were negative for ica genes. This can be due to low number of no biofilm producer isolates in present study. However, high rate of biofilm formation and high prevalence of ica genes can indicate importance of the presence of these genes in pathogenesis of this bacteria. Another study showed S. aureus strains, despite having the ica locus may fail to form biofilm in vitro as biofilm formation on inert surfaces is highly sensitive to growth conditions [27]. A previous study reported slime-positive S. aureus and S. epidermidis strains were deficient in the icaA and icaD genes as well as the whole ica locus. They suggested that the changed phenotype might be associated with the deletion of the entire ica locus [8].
There are several reports concerning prevalence rate of ica genes in S. aureus from different countries [7]. In the study on 63 MRSA clinical isolates, 29 (46%) of the isolates were shown to have strong ability to produce biofilm, and all the isolates carried icaD and icaC genes, whereas, the prevalence of icaA and icaB was 60.3% and 51%, respectively [22]. Comparatively, S. aureus isolates in the present study were not strong biofilm producers. In addition, S. aureus isolates from the urine and blood were not strong biofilm producers in comparison to those isolated from wound specimens. In another study by Hou W et al., among 55.56% of S. aureus isolates that produced biofilm phenotypically, 11.11% had icaA gene, but other genes were not investigated [28]. Compatible to other research findings we found MRSA isolates to harbour higher rate of icaADBC genes. However, Smith K et al., and Atshan SS et al., detected no significant correlation between susceptibility to methicillin and biofilm formation [29,30]. In the present study, among 18 MRSA isolates, only one isolate was strong producer, while four were moderate producers and 12 of them were found to be weakly adherent. While, one isolate did not form biofilm. On the other hand, among MSSA isolates, 4 (5.1%) isolates were found to be strong producers, 24 (30.4%) were moderate and 43 (54.4%) were found weakly adherent.
Limitation
The main limitation of the present study was presence of small number of non biofilm producing isolates. Future study should have larger number of isolates, includes non-biofilm producing isolates, to have better understanding and confirmation of the these results.
Conclusion
In conclusion, though there was a high prevalence of biofilm production among S. aureus isolated from inpatients specimens and majority of biofilm producing staphylococci isolates were positive for ica genes. Findings of the present study indicate importance and high rate of biofilm formation and the presence of ica genes family in pathogenic S. aureus. icaA and icaD were present in all MRSA isolates and all of these isolates were biofilm producer. There was no relation between presence of ica genes family and biofilm formation in our isolates. Controlling biofilm formation and use of ica genes for defining pathogenesis and control of infection can be an alternative therapies in future treatment.
ΨMRSA:Methicillin resistance S. aureus; MSSA:Methicillin sensitive S. aureus*Descriptive analysis and chi square tests were applied for analysis
ΨNot significance (because of absence of negative icaD isolate)
ΨNot significant (because of absence of vancomycin resistance isolate)