Introduction
Gastroenteritis is defined as the inflammation of the mucus membranes of the gastrointestinal tract and is characterized by diarrhoea or vomiting. Though it is a common and self-limiting childhood disease, it can lead to complications such as dehydration associated with electrolyte imbalance and acidosis, diminished growth, and impaired cognitive development particularly in resource-limited countries [1]. In developing countries, diarrhoea causes an estimated two million deaths annually among children aged <5 years [2].
Agents causing gastroenteritis include viruses, bacteria and protozoa. In children, 70% of cases of AGE are caused by viruses [1]. Four viral families are commonly associated with AGE: Reoviridae (Group A Rotaviruses), Caliciviridae (Noroviruses), Adenoviridae (Adenoviruses 40/41), and Astroviridae (Astroviruses), rarely Toroviruses, Picobirnaviruses, Picornavirus and Enterovirus 22 may also be associated [3].
Globally, Rotavirus AGE leads to 453,000 deaths and over two million hospitalizations among children under the age of five years [1]. In countries that have added Rotavirus vaccine to their routine childhood immunization policy the incidence and severity of Rotavirus infections have declined significantly [1,3]. Norovirus is now being recognized as most frequent cause of childhood gastroenteritis causing sporadic cases and outbreaks [1]. Each year Norovirus cause 64,000 episodes of diarrhoea requiring hospitalization and 900,000 clinic visits among children in industrialized countries, and up to 200,000 deaths of children <5 years of age in the developing countries [4].
Noroviruses are named after the original Norwalk strain, which caused an outbreak of AGE in a school in Norwalk, Ohio in 1968. Human Noroviruses belong to the family Caliciviridae, which is divided into four genera, Norovirus and Sapovirus, which cause human infections and Lagovirus and Vesivirus, which are associated with veterinary infections [5]. Noroviruses are small, single-stranded, nonenveloped, positive sense Ribonucleic Acid (RNA) viruses with a genomic size of approximately 7.5 kb [6]. Noroviruses are currently classified into five different genogroups (G) GI – GV, of which GI, II, and IV infect humans. GIII and GIV infect bovine and murine species, respectively [6].
Globally, Rotavirus is the predominant pathogen associated with gastroenteritis in children [1,7-9]. However, the prevalence of Norovirus in developing countries may be underestimated due to limitation of diagnostics tests available [10]. In India, there is limited data on Noroviruses associated diarrhoea in children [11]. The present study aims to evaluate pathogens associated with AGE in children of south Mumbai over a period of one month. Knowledge on the current epidemiology of the agents associated with AGE in the given geographic area will help in better management and timely practice of infection control measures.
Materials and Methods
A prospective study was carried out in July 2013 at a tertiary care hospital to investigate the cause of diarrhoea in children along with molecular testing at a reference center. Informed consent was obtained from parents of each child before inclusion in the study. This study was approved by the Hospital Ethics Committee. Children ≤ 12 years of age, having AGE were included in the study. Children with history of antibiotics treatment or hospitalization in the previous month were excluded from the study. diarrhoea was defined as the passage of three or more loose or liquid stools per day accompanied with or without vomiting and/or other symptoms such as fever, nausea, abdominal pain and cramps [12]. Clinical history was recorded in a case record form. Severity of disease was assessed by the modified Vesikari scoring pattern [13].
Stool samples were collected from both OPD and hospitalized children having diarrhoea in a wide mouth container provided by the hospital and was processed within one hour of sample receipt. Collected samples were processed for stool routine microscopy and culture as per hospital laboratory protocol. Gross examination of the stool samples included colour, consistency and reporting of occult blood. Routine microscopy was performed under 10x and 40x (Saline and Iodine preparation) for Red Blood Cells (RBCs), Pus cells, and Parasites (Ova and Cysts). For routine bacterial cultures MacConkey agar and Salmonella Shigella agar were inoculated and incubated aerobically at 35°C. Enrichment was done in Selenite F broth and after incubation for two hours at 35°C subculture was done on MacConkey agar and Salmonella Shigella agar. Identification and susceptibility testing of suspected bacterial pathogens was performed on Vitek 2 Compact (BioMérieux Pvt., Ltd.,).
A part of fresh stool sample was frozen immediately and stored at –20°C. Maintaining the cold chain, the stored samples were transported to reference centre in batches for molecular testing (PCR) for Rota Virus, Adenovirus, Norovirus and Enterovirus.
Molecular Testing for Viral Aetiology
RNA extraction and reverse transcription: Viral RNA was extracted from 30% (w/v) stool suspensions in Minimal Essential Medium (MEM) using TRIzol (Invitrogen, USA) according to manufacturer’s instructions and the viral RNA obtained was dissolved in RNase free water and stored at –20oC until used. A cDNA was synthesized at 37oC using random primers pd(N)6 (Roche, Diagnostics) and 100UM-MLV reverse transcriptase (Invitrogen).
Detection and characterization of Norovirus strains: Norovirus GI and GII were detected by PCR using cDNA as the template with primers Mon 432 and Mon 434 and primers Mon 431 and Mon 433 respectively [14]. The PCR amplificates (213 bp) were analysed on the 2% agarose gel.
Detection of Enteroviruses: The cDNA was used as the template for the detection of the 5’NCR of the Enterovirus gene by using panEV PCR primers EV1 and EV2 [15]. The amplification products were analysed on 10% polyacrylamide gels after staining with 0.5 μg/mL ethidium bromide to visualize PCR amplification bands using UV Transilluminator (BioRad).
Enterovirus Isolation: The stool samples were treated with chloroform before inoculating in the Rhabdomyosarcoma (RD) cells for virus isolation according to World Health Organization (WHO) protocol [16].
The cultures were incubated at 36oC and the Cytopathic Effect (CPE) as observed for five days. Samples not showing CPE were passaged again and the CPE was observed for another five days. If the CPE was observed, tissue culture material was harvested for RNA extraction for virus identification. Samples were scored negative when three serial passages did not produce CPE. Tubes showing CPE were harvested after two freeze-thawing cycles. Virus isolates were identified by partial VP1 sequencing as suggested by Oberste et al. [17].
Partial VP1 sequencing and Enterovirus serotype identification: The cDNA was used for partial VP1 PCR using 222/224 or AN88/ AN89 primer pairs [18]. The PCR products were used for Enterovirus serotype identification by sequencing on automated DNA sequencer ABI 3130xl using BigDye Terminator v3.1 ready reaction cycle sequencing kit (Applied Biosystem, Forster City, CA) as per the manufacturer’s instructions. The primers used for PCR amplification were used as the sequencing primers. The Enterovirus serotypes were identified by comparison of the VP1 sequences obtained with database of VP1 sequences of all EV serotypes from GenBank using BLASTn programme.
Detection of Rotavirus: The cDNA was used as template for the detection of Rotavirus using NSP4F and NSP4R PCR primers [19]. The Amplification products were analysed on 1% agarose gel.
Detection of Adenovirus: The cDNA was used as the template for the detection of Adenovirus using Ad40 and Ad41 PCR primers [20]. The Amplification products were analysed on 1% agarose gel.
Details of primers used are given in [Table/Fig-1] [14,15,18-20].
Primers used for PCR amplification and sequencing [14,15,18-20].
| Viruses | Primers | Sequences (5’-3’) | Region | Nucleotide positions | PCR product size (bp) |
|---|
| Norovirus [14] | Mon 432 | TGGACICGYGGICCYAAYCA | B (GI) | 5093 – 5112 | 213 bp |
| Mon 434 | GAASCGCATCCARCGGAACA | 5285 – 5305 |
| Mon 431 | TGGACIAGRGGICCYAAYCA | B (GII) | 5093 – 5112 | 213 bp |
| Mon 433 | GAAYCTCATCCAYCTGAACA | 5285 - 5305 |
| Enterovirus [15,18] | EV1 | ACACGGACACCCAAAGTAGTCGGTTCC | 5’NCR | 539-565 | 114 bp |
| EV2 | TCCGGCCCCTGAATGCGGCTAATCC | 5’NCR | 452-476 |
| 224 | GCIATGYTIGGIACICAYRT | VP3 | 1977-1996 | 762 bp |
| 222 | CICCIGGIGGIAYRWACAT | VP1 | 2969-2951 |
| AN89 | CAGCACTGACAGCAGYNGARAYNGG | VP1 | 2602-2627 | 372 bp |
| AN88 | TACTGGACCACCTGGNGGNAYRWACAT | VP1 | 2977-2951 |
| Rotavirus [19] | NSP4F | GGCTTTTAAAAGTTCTGTTCCG | NSP4F | 1-22 | 743 bp |
| NSP4R | GTCACACTAAGACCATTCC | NSP4R | 753-732 |
| Adenovirus [20] | Ad40 | GCCGCAGTGGTCTTACATGCACATC | Hexon | 18858-18882 | 300 bp |
| Ad41 | CAGCACGCCGCGGATGTCAAGT | Hexon | 19136-19158 |
Standard IUB nucleotide ambiguity codes are used:
I=Deoxyinosine; N=G, A, T or C; Y=C or T W=A or T: R=A or G
The locations of all primers are those relative to the genome of PV1 Mahoney (GenBank accession number J02281).
Results
Fifty-one children ≤12 years of age with AGE were included in the study. All the patients included in the study were vaccinated for Rotavirus. Of 51 patient tested, 45.10% (23/51) were positive for viruses by real time PCR. Routine stool culture was positive for only one patient for pathogenic E. coli O157. One patient had Entamoeba histolytica infection. Of 23 patients positive for viral aetiology, Norovirus were identified in 41.18% patients (21/51), Enterovirus was seen in 3.92% (2/51) patients and two patients had mixed infection with Norovirus, Enterovirus and Rotavirus [Table/Fig-2].
Virus aetiology and their Genotypes.
| Patients positive for Viral Aetiology | Norovirus (NV) Genogroup | Rotavirus (RV) | Enterovirus (EV) | Mixed Infection |
|---|
| GI | GII | GI + GII | RV+NoVGI+EV | NoVGI+GII +EV |
|---|
| 23 | 9 | 8 | 2 | 0 | 2 | 1 | 1 |
Our study group included 43 outpatients and 8 inpatients. Norovirus positivity was 41.86% (18/43) and 37.5% (3/8) among outpatients & inpatients respectively. Both the patients positive for Enterovirus were out patients.
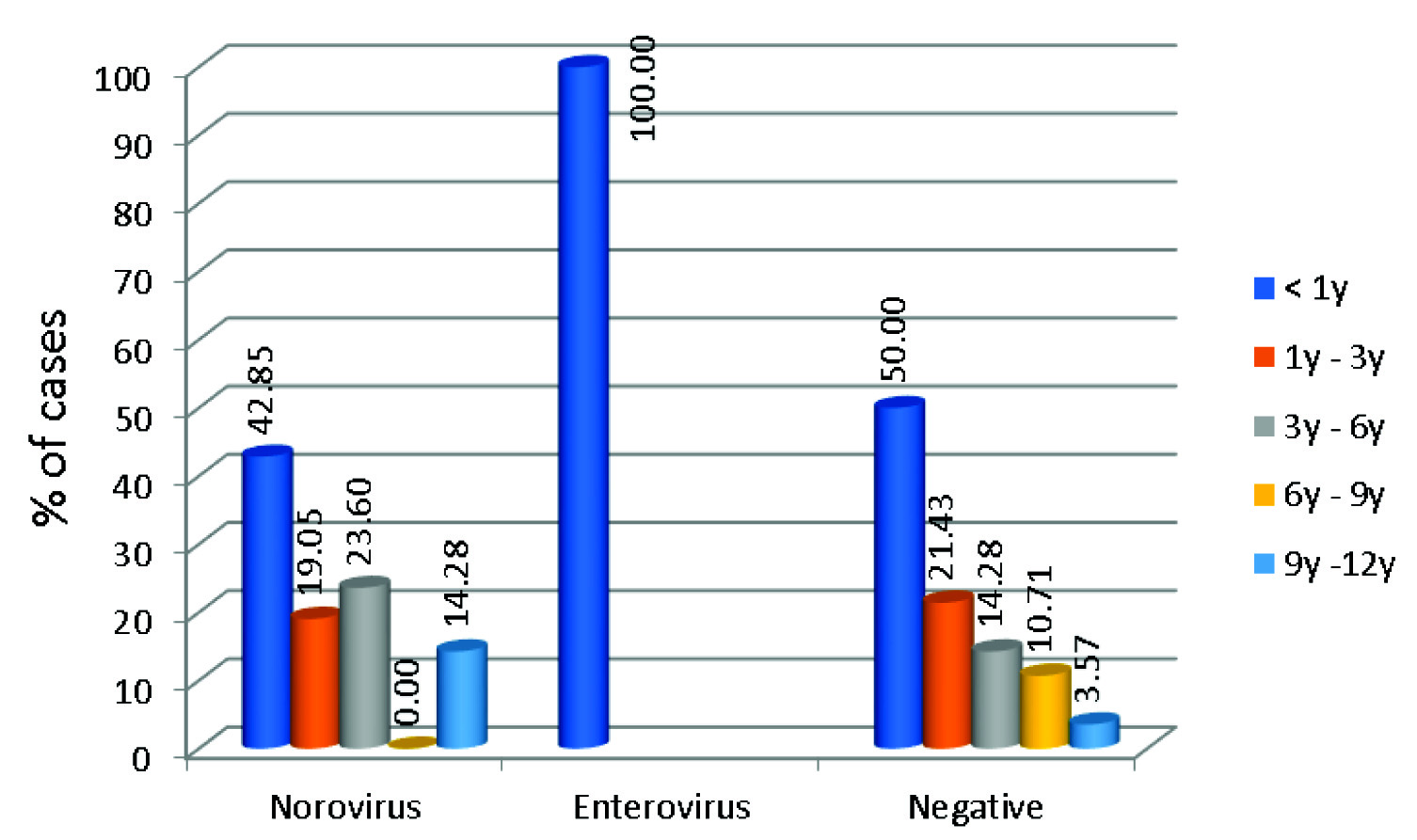
In our study, 65.21% (15/23) of children under three years of age had viral diarrhoea. Age distribution of patients who were Norovirus positive is given in [Table/Fig-3].
Agewise distribution of patients positive and negative for Viral aetiology.
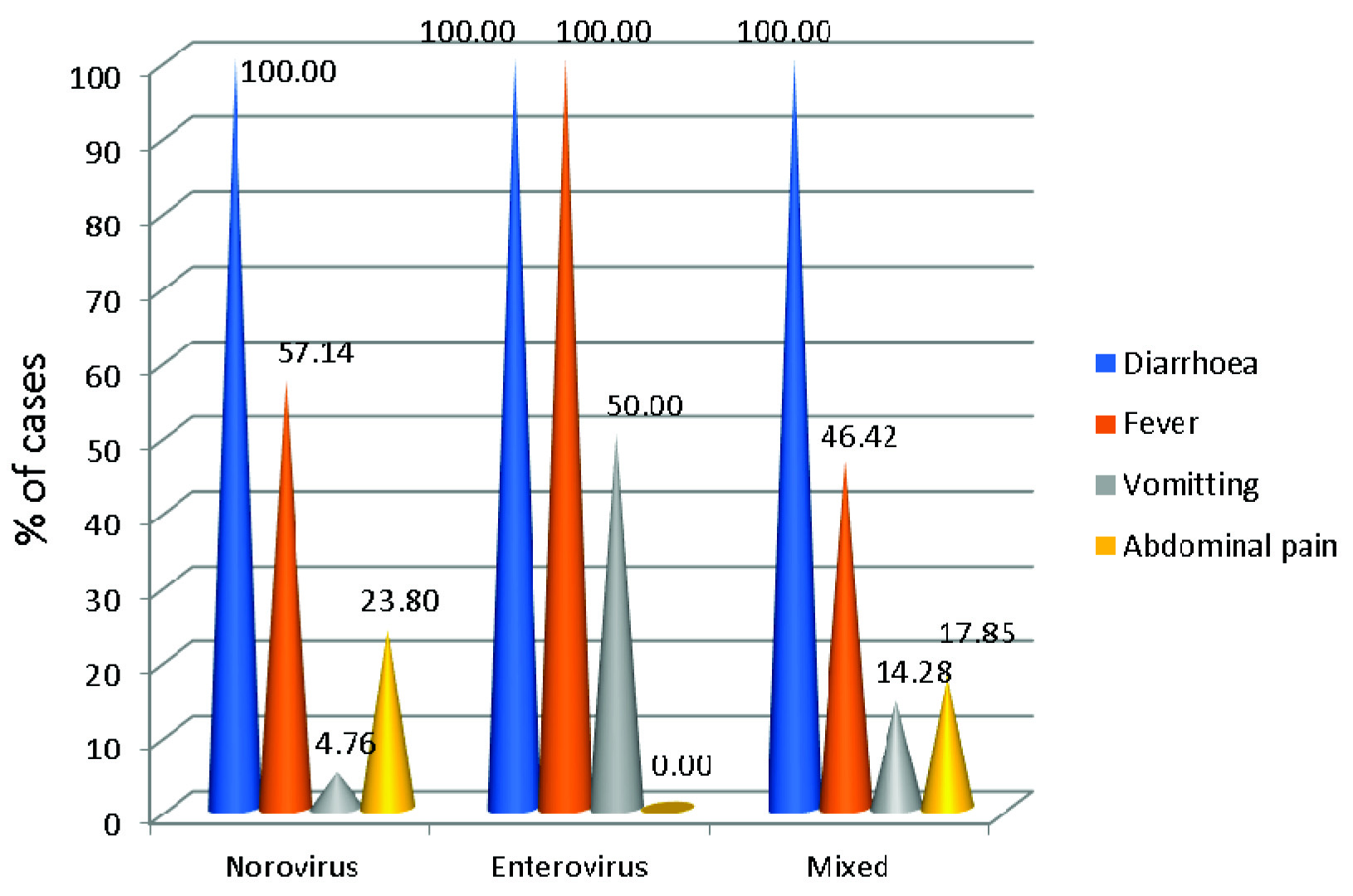
Overall diarrhoea was the most common presentation followed by fever in both those tested negative and those positive. Vomiting and abdominal pain were less common presentations [Table/Fig-4]. Clinical severity of disease was assessed by using modified Vesikari scoring system by (Ruuska and Vesikari (1990) [Table/Fig-5,6] [12].
Clinical symptoms of the patients included in the study.
Vesikari clinical severity scoring system severity rating scale and scores.
| Severity category |
| Mild | Moderate | Severe | Maximum score |
| <7 | 7 -10 | >=11 | 20 |
| Score |
| Parameter | 1 | 2 | 3 |
| Diarrhoea | |
| Maximum stools per day | 1-3 | 4-5 | >=6 |
| Diarrhoea duration (days) | 1-4 | 4 | >=6 |
| Vomiting | |
| Maximum number vomiting episodes per day | 1 | 2-4 | >=5 |
| Vomiting duration (days) | 1 | 2 | >=3 |
| Temperature | 37.1 -38.4 | 38.5 -38.9 | >=39 |
| Dehydration | N/A | 1 -5 % | >=6% |
| Treatment | Rehydration | Hospitalisation | N/A |
N/A: Not applicable
Lewis K. Vesikari Clinical Severity Scoring System manual. Seattle: PATH, 2011.
Discussion
In present study, Norovirus were identified in 41.18% patients (21/51). Chhabra P et al., reported a prevalence of 6.3% -12.6 % in children < 7yrs of age suffering from AGE from Western India [6]. In India, the prevalence varies from 6.6 to 25.7% from Southern and Northern India [8,21]. Some studies have reported lower prevalence of 1.4 % to 2.3% [22,23]. In Tunisia, the prevalence of Norovirus in sporadic cases was found to be 9.3% [24]. A prevalence of 25% was reported by Zeng M et al., in China [25].
Globally and in India, GII is the predominant genogroup causing gastroenteritis in all age groups [1,8,9]. Contrary to this, in our study we observed that genogroup I (47.61%) was predominant. Further molecular studies are needed to confirm this variation in the epidemiology observed in our study.
Global epidemiology of AGE indicates that Rotavirus is the predominant pathogen causing diarrhoea in children [1,8]. Contrary to this, in our study Norovirus was the predominant viral pathogen. Clinical history reveals all the patients included in our study were vaccinated against Rotavirus. Studies have shown that in the Rotavirus vaccine era, Norovirus is emerging as an important pathogen causing enteric infection in children [1,23]. Present study points out to this changing epidemiology of acute viral gastroenteritis.
Globally, the prevalence of mixed infections reported from various studies ranges from 4.4% to 14% [26-28]. Studies from India have shown mixed infections in the range of 0.6% to 23.2% [8,9,22], while in our study co-infections were seen in 3.92% of patients [Table/Fig-5]. Coinfection is commonly reported with Rotavirus and Norovirus or Adenovirus or Enterovirus or Astrovirus [8,9,27,28].
Severity of disease among study group
| Clinical severity score | Norovirus (n=21) | Enterovirus (n=2) | Negative (n=28) |
|---|
| <7 | MILD | 66.66 | 50 | 78.57 |
| 7-10 | MODERTAE | 33.33 | 50 | 14.28 |
| >11 | SEVERE | 0 | 0 | 7.14 |
In this study, Norovirus was detected in 41.86% (18/43) and 37.5% (3/8) outpatients and inpatients respectively. In a study from south India, 9.4% and 15.1% positivity were seen among outpatients & inpatients respectively [21]. Positivity among outpatients and hospitalized patients in western India was 9% and 12.5% respectively [11]. Gupta S et al., reported 2.3% of hospitalized patients positive for Norovirus [8]. Higher rates of 34% in hospitalized children similar to our study have been reported from Japan [29]. China and Italy report, 25.6% and 47% of hospitalized children with Norovirus infections respectively [25,30].
Although Norovirus were detected in all age groups, the infection with Norovirus was more commonly seen in children less than one year of age (42.85%). Chhabra P et al., in a study from western India also reports 40% of children ≤ one year of age to be infected by Norovirus [6]. In a study from Delhi and Himachal Pradesh Norovirus infection was seen in children < 2 year of age [8,22]. In China, it is seen in children <3 years of age [25]. In Tunisia, Norovirus was frequently seen in children more than 35 months [24].
Vomiting along with diarrhoea and fever is the most common presentation of Norovirus infection. Sai L et al., reported 67.5% children had vomiting and 46.3% had fever [31]. Similarly, in Taiwan, vomiting was reported as a more common presentation compared to fever and diarrhoea [32]. In our study, only 4.76% of children presented with vomiting. Study from Western India has reported 32% of hospitalized and 43% outpatients did not have vomiting [6]. Gupta S et al., reported increased in duration of vomiting for Norovirus infected patients, though not statistically significant (p-value=0.076) [8]. Study by Ramirez S et al., has also reported absence of vomiting in 51% of Norovirus infected patients [30].
Norovirus infection is called as “Winter Vomiting Disease” in West. In present study, maximum cases were observed during the monsoons. In Indian studies, Norovirus infection was reported in summer as well as winter season [6,8]. In Taiwan, higher incidence was seen in winter months [32] whereas in China, season of Norovirus associated disease varies with the region [25].
According to modified Vesikari scoring system, 66.66% of patients with Norovirus diarrhoea had mild disease and 33.33% had moderate disease and no cases of severe illness were reported. Chhabra P et al., reported, 29%, 57% and 14% of outpatients with severe, moderate and mild disease respectively [6]. In our study, 14% of Norovirus affected children required hospitalization. Contrary to this, Chhabra P et al., reported >50% hospitalized patients with Norovirus infection having severe disease [6]. Another study from western India reported 29.2% and 70.8% of the children having moderate and severe disease respectively [11].
Gonzalez-Galan V et al., in Spain reported 20% of patients with Norovirus infection had co-infection with Salmonella spp. [33]. In present study, one patient with Norovirus infection was co-infected with E. coli 0157.
Worldwide, Norovirus is the leading cause of non-bacterial gastroenteritis among individuals of all age groups [34]. Apart from symptomatic disease, Norovirus infection can lead to asymptomatic infections which serve as reservoirs of infection [21]. Norovirus has very low infecting dose and transmission takes place by multiple modes such as directly, from person to person; via contaminated food or water; via airborne droplets of vomitus and through contaminated environmental surfaces. Its ability to resist desiccation and disinfection favours its survival in the environment for long period of time allowing transmission via fomites. All these features add to its potential to cause outbreaks in institutional settings e.g., nursing homes, hospitals, military camps, cruise and schools [35,36]. Norovirus generally causes mild to moderate disease however severe disease is seen in the vulnerable population like children, elderly or immunocompromised patients [37,38]. There is no specific treatment available for Norovirus gastroenteritis, the therapy is only supportive [36].
Currently there are no licensed vaccines available for Norovirus [36,38]. Challenges associated with development of vaccine include inability to grow the virus in culture, genetic/antigenic diversity, limited knowledge on Norovirus immunology and assessment of vaccine performance in naïve individuals. NoV-Virus-Like Particles (VLPs) produced by recombinant technology have been identified as potential vaccine candidates and have shown good serum response [36,38,39]. But the success of a commercially available NoV vaccine, in developing countries will depend upon factors like cost, need for maintenance of a cold chain and integration into an already crowded immunization program.
Limitation
A limitation of study is the small size of the subjects as the study was conducted during a period of one month. Multicentric studies are needed to understand the epidemiology of the virus in India and to put measures for infection prevention and control, particularly in the era of Rotavirus vaccine.
Conclusion
In the era of Rotavirus vaccine, Norovirus could become an important pathogen causing AGE. As Norovirus disease is difficult to distinguish clinically from other causes of AGE, a syndromic approach based testing of AGE may be considered in the diagnostic algorithm.
Standard IUB nucleotide ambiguity codes are used:I=Deoxyinosine; N=G, A, T or C; Y=C or T W=A or T: R=A or GThe locations of all primers are those relative to the genome of PV1 Mahoney (GenBank accession number J02281).
N/A: Not applicableLewis K. Vesikari Clinical Severity Scoring System manual. Seattle: PATH, 2011.
[1]. El Qazoui M, Oumzil H, Baassi L, El Omari N, Sadki K, Amzazi S, Rotavirus and norovirus infections among acute gastroenteritis children in MoroccoBMC Infect Dis 2014 14(1):30010.1186/1471-2334-14-30024894194 [Google Scholar] [CrossRef] [PubMed]
[2]. Centers for Disease Control and Prevention. Managing acute gastroenteritis among children: oral rehydration, maintenance, and nutritional therapy. Atlanta, GA: Centers for Disease Control and Prevention – Federal Government Agency [U.S.]. 2003 Nov 21. Available from: https://www.cdc.gov/mmwr/pdf/rr/rr5216.pdf Accessed on July 18,2017 [Google Scholar]
[3]. Glass RI, Bresee J, Jiang B, Gentsch J, Ando T, Fankhauser R, Gastroenteritis viruses: an overviewNovartis Found Symp 2001 238:5-25.10.1002/0470846534.ch211444035 [Google Scholar] [CrossRef] [PubMed]
[4]. Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD, Systematic literature review of role of noroviruses in sporadic gastroenteritisEmerg Infect Diseases 2008 14(8):122410.3201/eid1408.07111418680645 [Google Scholar] [CrossRef] [PubMed]
[5]. Lindell AT, Grillner L, Svensson L, Wirgart BZ, Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in childrenJ Clin Microbiol 2005 43(3):1086-92.10.1128/JCM.43.3.1086-1092.200515750066 [Google Scholar] [CrossRef] [PubMed]
[6]. Chhabra P, Dhongade RK, Kalrao VR, Bavdekar AR, Chitambar SD, Epidemiological, clinical, and molecular features of norovirus infections in western IndiaJ Med Virol 2009 81(5):922-32.10.1002/jmv.2145819319938 [Google Scholar] [CrossRef] [PubMed]
[7]. Nair GB, Ramamurthy T, Bhattacharya MK, Krishnan T, Ganguly S, Saha DR, Emerging trends in the aetiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, IndiaGut Pathog 2010 2:410.1186/1757-4749-2-420525383 [Google Scholar] [CrossRef] [PubMed]
[8]. Gupta S, Krishnan A, Sharma S, Kumar P, Aneja S, Ray P, Changing pattern of prevalence, genetic diversity, and mixed infections of viruses associated with acute gastroenteritis in pediatric patients in New Delhi, IndiaJ Med Virol 2018 90:469-76.10.1002/jmv.2498029064572 [Google Scholar] [CrossRef] [PubMed]
[9]. Chitambar S, Gopalkrishna V, Chhabra P, Patil P, Verma H, Lahon A, Diversity in the enteric viruses detected in outbreaks of gastroenteritis from Mumbai, Western IndiaInt J Environ Res Public Health 2012 9:895-915.10.3390/ijerph903089522690171 [Google Scholar] [CrossRef] [PubMed]
[10]. Koo HL, Ajami NJ, Jiang ZD, Neill FH, Atmar RL, Ericsson CD, Noroviruses as a cause of diarrhea in travelers to Guatemala, India, and MexicoJ Clin Microbiol 2010 48(5):1673-76.10.1128/JCM.02072-0920305012 [Google Scholar] [CrossRef] [PubMed]
[11]. Chhabra P, Chitambar SD, Norovirus genotype IIb associated acute gastroenteritis in IndiaJ Clin Virol 2008 42(4):429-32.10.1016/j.jcv.2008.03.01418467163 [Google Scholar] [CrossRef] [PubMed]
[12]. Thapar N, Sanderson IR, Diarrhoea in children: an interface between developing and developed countriesLancet 2004 363(9409):641-53.10.1016/S0140-6736(04)15599-2 [Google Scholar] [CrossRef]
[13]. Lewis K, Vesikari Clinical Severity Scoring System manual. Seattle: PATH, 2011Available from: https://www.path.org/publications/files/VAD_vesikari_scoring_manual.pdf Accessed on July 18,2017 [Google Scholar]
[14]. Ando T, Monroe SS, Gentsch JR, Jin QI, Lewis DC, Glass RI, Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and southern hybridizationJ Clin Microbiol 1995 33(1):64-71. [Google Scholar]
[15]. Chen-Fu Y, De L, Su-Ju Y, Gómez JR, Cruz J, Holloway BP, Genotype-specific in vitro amplification of sequences of the wild type 3 polioviruses from Mexico and GuatemalaVirus Res 1992 24(3):277-96.10.1016/0168-1702(92)90124-R [Google Scholar] [CrossRef]
[16]. WHO (2004). Polio Laboratory Manual. Geneva: World Health Organization [Google Scholar]
[17]. Oberste MS, Maher K, Kilpatrick DR, Flemister MR, Brown BA, Pallansch MA, Typing of human enteroviruses by partial sequencing of VP1J Clin Microbiol 1999 37(5):1288-93. [Google Scholar]
[18]. Nix WA, Oberste MS, Pallansch MA, Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimensJ Clin Microbiol 2006 44(8):2698-704.10.1128/JCM.00542-0616891480 [Google Scholar] [CrossRef] [PubMed]
[19]. Ciarlet M, Liprandi F, Conner ME, Estes MK, Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotavirusesArch Virol 2000 145(2):371-83.10.1007/s00705005002910752559 [Google Scholar] [CrossRef] [PubMed]
[20]. Allard A, Albinsson B, Wadell G, Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reactionJ Med Virol 1992 37(2):149-57.10.1002/jmv.18903702141629713 [Google Scholar] [CrossRef] [PubMed]
[21]. Monica B, Ramani S, Banerjee I, Primrose B, Iturriza-Gomara M, Gallimore CI, Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South IndiaJ Med Virol 2007 79(5):544-51.10.1002/jmv.2086217385696 [Google Scholar] [CrossRef] [PubMed]
[22]. Jain S, Thakur N, Grover N, Vashistt J, Changotra H, Prevalence of rotavirus, norovirus and enterovirus in diarrheal diseases in Himachal Pradesh, IndiaVirus Dis 2016 27:77-83.10.1007/s13337-016-0303-226925447 [Google Scholar] [CrossRef] [PubMed]
[23]. Gupta S, Singh KP, Jain A, Srivastava S, Kumar V, Singh M, Aetiology of childhood viral gastroenteritis in Lucknow, north IndiaIndian J Med Res 2015 141:469-72.10.4103/0971-5916.15929826112849 [Google Scholar] [CrossRef] [PubMed]
[24]. Hassine-Zaafrane M, Sdiri-Loulizi K, Kaplon J, Salem IB, Pothier P, Aouni M, Prevalence and genetic diversity of norovirus infection in Tunisian children (2007-2010)J Med Virol 2013 85(6):1100-10.10.1002/jmv.2355223532785 [Google Scholar] [CrossRef] [PubMed]
[25]. Zeng M, Xu X, Zhu C, Chen J, Zhu Q, Lin S, Clinical and molecular epidemiology of norovirus infection in childhood diarrhea in ChinaJ Med Virol 2012 84(1):145-51.10.1002/jmv.2224822028199 [Google Scholar] [CrossRef] [PubMed]
[26]. Levidiotou S, Gartzonika C, Papaventsis D, Christaki C, Priavali E, Zotos N, Viral agents of acute gastroenteritis in hospitalized children in GreeceClin Microbiol Infect 2009 15(6):596-98.10.1111/j.1469-0691.2009.02855.x19604279 [Google Scholar] [CrossRef] [PubMed]
[27]. HamidReza MS, AliReza SZ, Manoochehr M, GholamReza K, Ahmad SZ, Shahram J, Astrovirus and rotavirus co-infections in children with gastroenteritis who were referred to Ahvaz Aboozar Hospital, Southern IranJundishapur J Microbiol 2012 5(1):352-54. [Google Scholar]
[28]. de Oliveira Ferreira CE, Raboni SM, Pereira LA, Nogueira MB, Vidal LR, Kashyap LR, Almeida SM, Viral acute gastroenteritis: clinical and epidemiological features of co-infected patientsBraz J Infect Dis 2012 16(3):267-72.10.1016/S1413-8670(12)70322-7 [Google Scholar] [CrossRef]
[29]. Kawada JI, Arai N, Nishimura N, Suzuki M, Ohta R, Ozaki T, Clinical characteristics of norovirus gastroenteritis among hospitalized children in JapanMicrobiol Immunol 2012 56(11):756-59.10.1111/j.1348-0421.2012.00498.x22889384 [Google Scholar] [CrossRef] [PubMed]
[30]. Ramirez S, De Grazia S, Giammanco GM, Milici M, Colomba C, Ruggeri FM, Detection of the norovirus variants GGII. 4 hunter and GGIIb/hilversum in Italian children with gastroenteritisJ Med Virol 2006 78(12):1656-62.10.1002/jmv.2075117063517 [Google Scholar] [CrossRef] [PubMed]
[31]. Sai L, Sun J, Shao L, Chen S, Liu H, Ma L, Epidemiology and clinical features of rotavirus and norovirus infection among children in Ji’nan, ChinaVirol J 2013 10(1):30210.1186/1743-422X-10-30224099150 [Google Scholar] [CrossRef] [PubMed]
[32]. Wu TC, Liu HH, Chen YJ, Tang RB, Hwang BT, Yuan HC, Comparison of clinical features of childhood norovirus and rotavirus gastroenteritis in TaiwanJ Chin Med Assoc 2008 71(11):566-70.10.1016/S1726-4901(08)70170-9 [Google Scholar] [CrossRef]
[33]. Gonzalez-Galan V, Sánchez-Fauqier A, Obando I, Montero V, Fernandez M, Torres MJ, High prevalence of community-acquired norovirus gastroenteritis among hospitalized children: a prospective studyClin Microbiol Infect 2011 17(12):1895-99.10.1111/j.1469-0691.2011.03506.x21848976 [Google Scholar] [CrossRef] [PubMed]
[34]. Menon VK, George S, Ramani S, Illiayaraja J, Sarkar R, Jana AK, Genogroup IIb norovirus infections and association with enteric symptoms in a neonatal nursery in southern IndiaJ Clin Microbiol 2010 48(9):3212-15.10.1128/JCM.02510-0920631107 [Google Scholar] [CrossRef] [PubMed]
[35]. Widdowson MA, Bulens SN, Beard RS, Lane KM, Monroe SS, Lance S, Enhanced surveillance of norovirus outbreaks of gastroenteritis in GeorgiaPublic Health Rep 2011 126(2):251-58.10.1177/00333549111260021621387955 [Google Scholar] [CrossRef] [PubMed]
[36]. Simons MP, Pike BL, Hulseberg CE, Prouty MG, Swierczewski BE, Norovirus: new developments and implications for travelers’ diarrheaTrop Dis Travel Med Vaccines 2016 2(1):110.1186/s40794-016-0017-x28883945 [Google Scholar] [CrossRef] [PubMed]
[37]. Desai R, Hembree CD, Handel A, Matthews JE, Dickey BW, McDonald S, Severe outcomes are associated with genogroup 2 genotype 4 norovirus outbreaks: a systematic literature reviewClin Infect Dis 2012 55(2):189-93.10.1093/cid/cis37222491335 [Google Scholar] [CrossRef] [PubMed]
[38]. Riddle MS, Walker RI, Status of vaccine research and development for norovirusVaccine 2016 34(26):2895-99.10.1016/j.vaccine.2016.03.07727036510 [Google Scholar] [CrossRef] [PubMed]
[39]. Lucero Y, Vidal R, O’Ryan GM, Norovirus vaccines under developmentVaccine 2017 S0264-410X(17):30830-37. [Google Scholar]