Introduction
The HBV is a major cause of liver disease in India and globally, has the second largest pool of CHB virus infections, a leading cause of HCC [1]. About 350 million people suffer from CHB and account for 55% of HCC cases worldwide. The fifth most common cause of cancer is HCC, and its incidence is increasing worldwide because of the dissemination of HBV infections [2]. While 95% of HBV infections in adults are resolvable acute infection, the remaining 5-10% develops CHB infection [3].
The HBV genome consists of a partially double-stranded circular DNA, approximately 3200 bp in length, with four overlapping open reading frames (ORFs: PreS1/PreS2/S, PreC/C, P and X) encoding seven polypeptides. HBV-X, the smallest gene, expresses a 154 amino acid multifunctional protein (HBx) with transcriptional transactivator activities. It has been suggested in prospective cohort studies that certain mutations in HBV-X may be associated with the development of HCC [4,5].
Mutational modifications of HBV-X may induce amino acid changes in the pol gene and pre C gene as their coding sequences overlap. Nucleotide mutations in the HBV-X/pre C region reportedly associated with HCC include the BCP bi-mutation, A1762T and G1764A which results in an HBeAg suppressive HBV phenotype [6]. Chronic Korean HBV genotype C2 carriers have eight key mutations: G1613A, C1653T, T1753V, A1762T, G1764A, A1846T, G1896A and G1899A throughout the core BCP and the proximal portion of the precore gene (X/preC region) that increase the risk for HCC [7]. Contradictory data on the association between these mutations and progression to HCC may be attributed to differences in the patient population, genotypes, HBeAg status and clinical stage of the patient [5].
Phylogenetic analyses of full-length HBV genome from various geographical regions of the world have classified HBV into nine genotypes (A-I). Specific genotypes are known to influence the course of the disease and prognosis of treatment. Genotypes A and D isolated from patients with liver disease had subgenotype A1 predominating in HCC patients from Southern India. A high prevalence of subgenotype A1 was seen among HCC patients. Irrespective of the infecting genotype, mutations A1762T/G1764A were significantly higher in HCC patients [8].
Kerala is one of the most densely populated state in India. HBsAg prevalence in north Kerala and among voluntary blood donors in South Kerala is reportedly 0.5% and 1.5% respectively. The present study investigated genetic variability of HBV-X in CHB patients from Central Kerala, India.
Materials and Methods
Study Population
In the present cross-sectional study a total of 15,002 blood samples were received from out-patient and in-patient wards of the Pushpagiri Institute of Medical Science and Research Centre (PIMS & RC), Tiruvalla, Kerala, India for HBsAg testing during January 2014 to March 2016. Serological prevalence of HBsAg positive subjects was 1.69% (n=255). Among these, CHBV patients (n=137) attending the Gastroenterology Unit with a history of HBsAg positivity >6 months were identified. Of the CHBV patients, 65.7% (n=90) were positive for HBV DNA (Core gene) and were included in this study. Patients who had the following complications were excluded from the study: Patients with alcoholic hepatitis, drug-induced hepatitis, other acute or chronic inflammatory diseases.
All patient details including sex, age, date of HBV diagnosis, biochemical tests and other investigations data were extracted from the medical records of study subjects. The study was approved by the Institutional Ethical Committee (ref: PIMSRC/E1/388A/37/2014).
DNA Extraction, Amplification and Genotyping of HBV
DNA was extracted from serum samples using QIAmp DNA Mini Kit (Qiagen Inc., Germany) and the core gene of HBV genome was amplified using primers as described previously by Kaneko S et al., [9]. A nested PCR for amplification of HBV-X was also done [4]. PCR products were run on 2% agarose gel, stained with SYBR Safe DNA gel stain (Invitrogen by Life Technologies, USA) and analysed. HBV genotyped were performed using multiplex PCR described by Naito H et al., [10]. All samples which could not identify were subjected to heminested PCR by the method of Krischberg O et al., [11].
Sequencing and Phylogenetic Analysis of HBV-X Sequences
Amplified products of HBV-X were sequenced using forward (5′-GCCGATCCATACTGCGGAACTC-3′) and reverse (5′-CCAAGGCACAGCTTGGAGGCT-3′) primers [4]. Phylogenetic analyses were done using MEGA version 7.0.21. All sequencing data were assembled, aligned and compared to consensus HBV reference sequences for each genotype and subgenotypes available at Genbank using ClustalW software (BioEdit version 7.2). Phylogenetic analysis was performed using maximum Likelihood tree method and bootstrap resampling was performed 1,000 times. Nucleotide sequences in the present study were deposited in GeneBank accession numbers: KX423692-KX423724, KX608922-KX608924, KX639490, KX646744 and KX712265.
Analysis of Mutations in HBV-X gene
Mutation analysis was performed in HBV-X using MEGA version 117.0.21 software by comparing with HBV reference sequences. The Kyte and Doolittle (1982) method, implemented in Bioedit version 7.2 was used to create hydrophobicity profile to identify potential antigenicity at genotype specific positions [12]. Non-synonymous (dN) and synonymous (dS) between the consensus sequences and signature amino acid patterns were calculated using sequence analysis tools: SNAP and VESPA, respectively using the program available at the Los Alamos National Laboratory website (http://www.hiv.lanl.gov).
Statistical Analysis
Results were represented as mean±SD, or as median (Interquartile range). Differences between continuous variables were compared using Student’s t-test. Chi-square test and Fisher’s exact test were used for categorical variables. For analysis of baseline data Microsoft office Excel 2007 (Microsoft Corporation, USA) was used. For statistical calculations, SPSS, version 12.0 was used with a statistically significant p-value <0.05.
Results
Baseline Characteristics of Study Population
The study consists of 90 HBV core DNA positive CHBV cases with known viral serological and biochemical parameters. Among the study population, 61 (67.78%) were male and 29 (32.23%) were female. The mean ages of male and female were highly significant, 55.94 (SD±15.77) and 49.46 (SD±17.24) years respectively (p<0.001). The mean ALT and AST were 50.96 (SD±50.13) IU/mL and 40.66 (SD±37.27) IU/mL. It was observed that there was a significant difference in ALT and AST levels between HBeAg-negative and positives (p=0.0186, p=0.0151, respectively). HBV X gene was amplified only from 39 (43.34%). Among this HBV X gene positive, 31 (79.48%) samples were positive for HBeAg.
Genotyping of HBV
Of the 90 HBsAg CHBV cases, 79 (87.78%) were successfully genotyped using the method described by Naito H et al., and Krischberg O et al., [10,11]. In the present study HBV genotype A was most prevalent strains (n=52, 57.77%) followed by genotype D (n=27, 30.00%). No other genotypes were observed in the present study. In genotype D patients prevalence of HBeAg was 81.48% (n=22), whereas only 50% (n=26) was observed in genotype A. The genotyping could not be determined for 11 (12.22%) strains, might have sequence variations which couldn’t amplify the target gene.
Phylogenetic Analysis of HBV-X Sequences
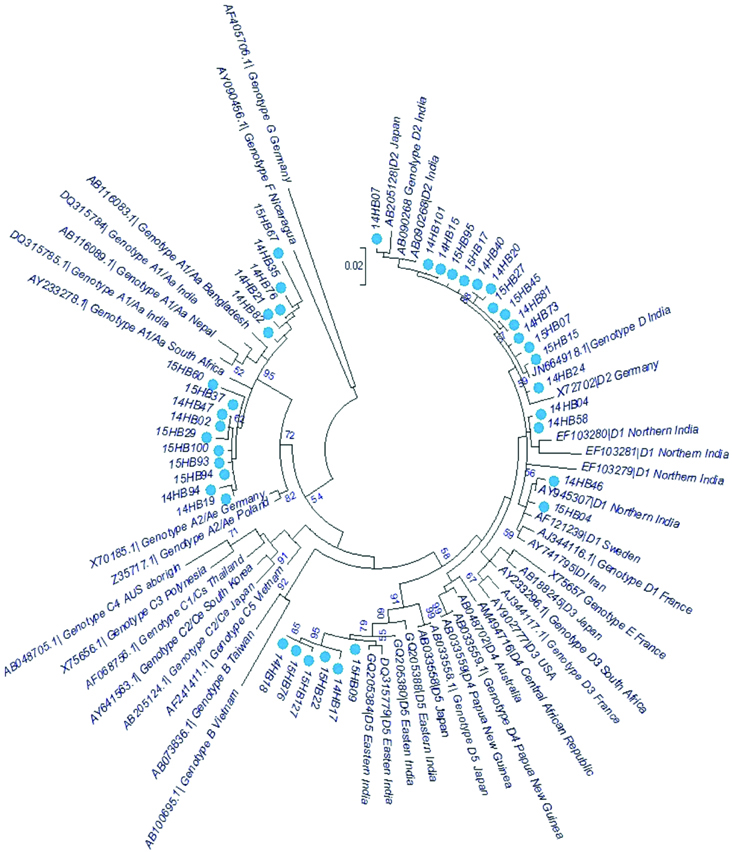
Phylogenetic analyses were performed to assess circulating genotypes and to examine heterogeneity of HBV-X in the present study group. Phylogenetic analysis of the HBV-X sequences along with GenBank reference sequences showed seven distinct clusters corresponding to the HBV genotypes (A-D and F-H) [Table/Fig-1]. Sequences segregated into two distinct clusters corresponding to HBV genotypes A (38.46 %, n=15) and D (61.53%, n=24). All genotype A sequences were ascribed to subgenotype A1. The A1 subgenotypes clustered within the Asian clade as a separate monophyletic clade with a common evolutionary ancestor (n=15). Genotype D sequences presented three monophyletic groups corresponding to subgenotype D2 (n=14), D5 (n=6) and D1 (n=4). In summary, the phylogenetic analysis showed D was most prevalent genotype in this study, followed by A.
Phylogenetic relationships among complete X gene sequences of Hepatitis B virus strains isolated from human (taxa name starting with ‘14HB’ or ‘15HB’, marked with the coloured round symbol) compared with sequences obtained from GenBank.
Mutation Analysis of HBV-X
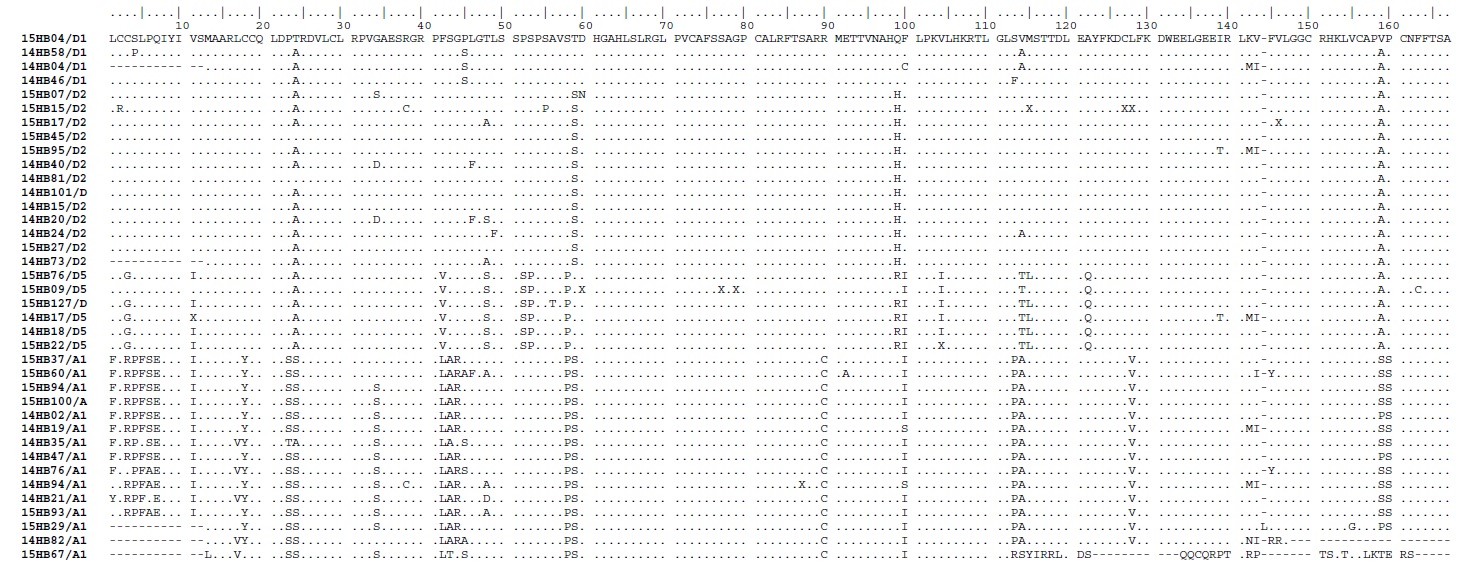
We detected double mutations (A1762T/G1764A) in 12.82% (5/39) sequences: mutations which have significantly been related to HCC in earlier studies [6,7]. Among these five double mutations strains, three belonged to D and two belonged to A genotypes. The double mutation was observed mainly in HBeAg negative patients (n=3). Among all the analysed sequences (n=39), three sequences had nucleotide substitutions (insertions) compared to the consensus reference sequences and were observed in two subgenotype D2 sequences (at positions1673-1703nt and 1675-1698nt) and in one A1 (at positions 1768-1772nt). These insertions caused a frameshift in the reading frame of nucleotide and thereby in the amino acid alignment. The A1 subgenotype sequences displayed the significantly higher number of nucleotide mutations compared to other subgenotypes in this study (p<0.0001). There was no significant association between patients age and genotypes (p=0.92). Alignment of the amino acid sequences is presented in and important mutations are summarised in [Table/Fig-2,3].
The amino acid sequences deduced from HBV-X nucleotides among the different genotypes within the study group.
A summary of nucleotide substitutions relevant to HBx mediated pathogenesis.
| S No. | Genotype | Age (Mean±SD) | HBeAg status | Number of samples with nucleotide mutation position |
|---|
| | | | C1484T | G1613A | T1740C | A1762T | G1764A | T1753C | C1495A |
|---|
| 1 | A1 (n=15) | 55.00±16.40 | Positive (n=11) | 11 | 1 | 7 | 1 | 2 | - | - |
| | | Negative (n=4) | 4 | 1 | 3 | 1 | 1 | - | - |
| 2 | D1 (n=4) | 58.80±24.10 | Positive (n=3) | - | - | - | - | - | - | - |
| | | Negative (n=1) | - | - | - | 1 | 1 | 1 | - |
| 3 | D2 (n=14) | 60.20±18.90 | Positive (n=11) | - | - | - | - | - | - | 1 |
| | | Negative (n=3) | - | - | - | 1 | 1 | - | - |
| 4 | D5 (n=6) | 60.80±10.70 | Positive (n=5) | - | - | - | 1 | 1 | 1 | - |
| | | Negative (n=1) | - | - | - | - | - | - | - |
Bioinformatic Analysis of HBV-X
Among the 39 HBV-X sequences most of the specific amino acid residues were located in the first 50 amino acid (N-terminal region) compared to the C-terminal regions. The genotype-specific amino acid mutations at 6, 11, 12, 22, 30, 31, 32, 46 and 47 were found in A1 genotypes and residues at 40, 41 and 46 were found in D5 genotypes, rest of the amino acid residues were well conserved within N-terminal domain sequences. At the C-terminus, residues at 78, 88, 109, 110 and 124 were shown to be A1 genotype dependent and residues at 87, 88, 92 and 155 were found to specify the D5 genotypes [Table/Fig-2].
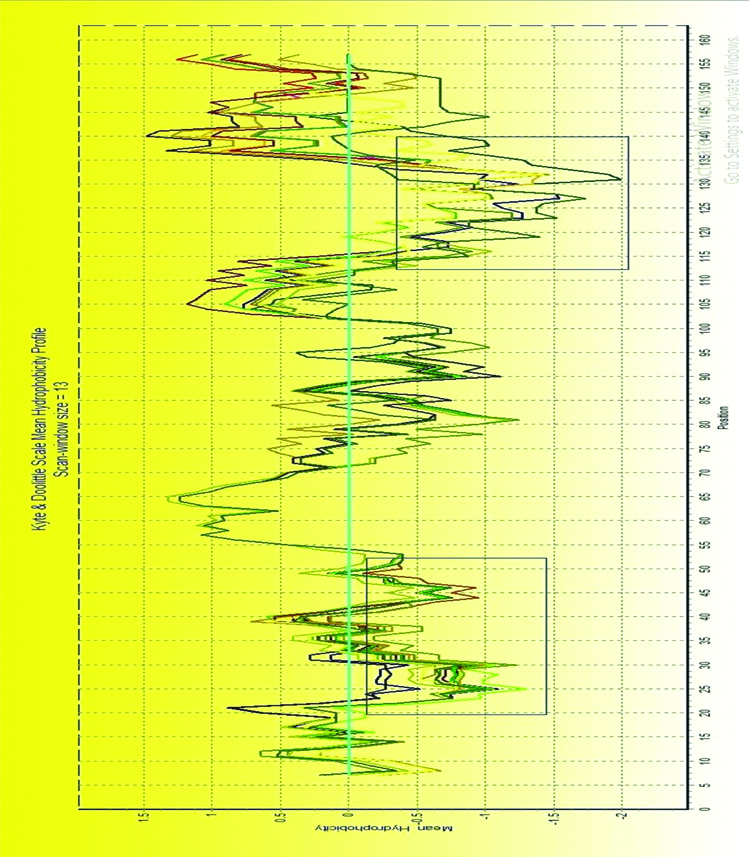
In Kyte-Doo-little Hydropathy Plots, hydropathy score represented the hydrophobic/hydrophilic character of amino acid. The higher the score, the greater the hydrophobicity is, the lower the score, the greater is the hydrophilic nature. As shown in [Table/Fig-4], these mutated positions were located in two regions of special hydrophobic/hydrophilic character, neutral area around position 35 and the hydrophilic area around position 110. The result indicated that mutations mainly affected the hydrophobic/hydrophilic character of HBV-X and may alter the pathology feature of HBV-X.
Kyte and Doolittle mean hydrophobicity profile of 39 aligned HBV-X aminoacid sequences.
Discussion
The HCC is the fifth most common cause of cancer in humans worldwide, especially in Asia and Africa [2]. Indian patients with CHB have a high risk of developing HCC. Various studies have reported that the clinical course of chronic HBV infection may be modified by specific mutation of HBV-X gene, although the significance of these mutations in patients with chronic hepatitis B infection remains controversial [13-15]. HBV-X gene translates a multifunctional protein which activates transcriptional transactivation and mediates cell growth via proliferation and apoptosis [14]. The nature of the HBV mutation may depend on patient’s age, sex, HBeAg status and viral load [15]. The present study focused on examining the entire HBV-X to identify specific known mutations. In this study, we observed that the number of male subjects was twice the number of females which corroborated with several studies from Asia and the world have demonstrated the predominance of chronic or active HBV infection among males [13,15].
Molecular testing was done for HBV core gene and HBV-X. The discrepancy between detection of core and HBV-X genes was high. Of the 90 HBV core gene study subjects, HBV-X could be amplified from 39 (43.34%) samples, suggesting the absence of X gene in the rest. Loss of HBV-X genetic region appears to be a unique feature of HBV strains circulating in our population. This may be due to splicing or HBV DNA being integrated into human chromosomes [16,17].
Genotype A was found to be the most prevalent in Central Kerala followed by Genotype D. Our results were similar to the previous reports from Kerala [18,19]. The results concur with early studies reported from Thiruvananthapuram, Kerala showed that A1 sub-genotype was the most prevalent and has a significant association with HCC [8]. Subgenotype A1 strains mainly clustered with the Asian strains. The patients belonging to genotype A and D did not show any significant difference in terms of age, HBeAg result and liver function parameters (AST and ALT).
Phylogenetic analysis of HBV-X gene sequences indicates similar genotype results that of the surface gene. Previous studies report that genotype effect in the chronic hepatitis B infection prognosis especially in genotype A, which more often results in chronic acute exacerbation with low HBV DNA levels [15]. A long-term further investigation needs to be done to clearly understand the influence of genotype contribution to the multi-step hepatocarcinogenesis process.
The HBV-X has been implicated in several liver diseases such as chronic hepatitis B and HCC [20]. HBV-X N and C-terminals act as a negative control of proapoptotic activity and promote transactivation, respectively [4,20]. The C1484T substitution was found only in sequences which were belonging to genotype A1 and suggesting that it may be genotype specific [4]. The present sequences had I127T due to T1753C mutation which appeared in two strains which clustered with D genotype. The I127T substitution has been shown to promote transactivation and increase anti-proliferative activity in cell culture [21]. It has been suggested that these can be due to HBV mechanism to evade the immune system leading to persistent viral replication which is important for hepatocarcinogenesis [22]. We observed that basal core promoter region mutations, namely, A1762T and G1764A were found mainly in HBeAg negative samples. These double mutations cause substitutions of K130M and V131I in the X protein. The bi-mutation A1762T/G1764A has been extensively studied by many groups and has been suggested to interfere with cell growth control, DNA repair, HBeAg synthesis and a strong inhibitor of p21 [19-21]. Patients with A1762T/G1764A dual mutations can be at risk of developing HCC and cirrhosis as the mutations are involved in multiple pathogenic effects.
The bioinformatics analyses identify most of HBX substitutions and mutations at hydrophilic sites. These mutations and substitutions changes may cause structural changes and conformation alteration of X protein and hence, alter the regulatory and transactivational functions [4,23]. However, further experiments need to be done to confirm the inference. This also suggests that the substitutions and mutations were HBV genotypes/subgenotypes specific.
Limitation
Follow-up with patients was done only for a year hence mutations could not be linked to carcinogenesis.
Conclusion
The HBV-X substitutions which circulate in different genotypes may affect the pathogenic potential of HBV due to complex interactions of the virus with the host. In the present study, genotype A1 isolates displayed most mutations/substitutions in the HBV-X. The pattern of the mutations formed may serve as prognostic factors as they represent a strategy of HBV to escape the immune system and lead to persistent liver infections which are important for hepatocarcinogenesis. This study represents the first formal investigation of HBV-X genetic variability in Kerala. However, a larger cohort would also be needed to validate the generalisation of the present results.
Statement of Author Contributions
Ozhiparambil A Jagan: Contributed to the design of study, carried out the laboratory testing, phylogenetic and mutation analysis, sequence submission, data analysis and drafted the manuscript; Gopi Manoj: Contributed to the design of study, data analysis, helped frame and edited the manuscript; Natamai S Jayaprakash: Contributed to the design of study, data analysis and reviewed the manuscript; Ramesh M Nair: Contributed to the design of study, clinically analysed the patients for inclusion into study, collected and sent samples for testing, reviewed the manuscript; Sara Chandy: Contributed to the design of study, supervised and defined intellectual content, helped in data acquisition and analysis, manuscript preparation and editing. The corresponding author of the manuscript.
All authors read and approved the final manuscript. Sara Chandy is the guarantor of the paper.
Conflict of Interest
The authors declare no conflict of interest or commercial affiliation related to this study.
[1]. Acharya SK, Madan K, Dattagupta S, Panda SK, Viral hepatitis in IndiaNatl Med J India 2006 19(4):203-17. [Google Scholar]
[2]. Kew MC, Hepatitis B virus x protein in the pathogenesis of hepatitis B virusinduced hepatocellular carcinomaJ Gastroenterol Hepatol 2011 26:144-52.10.1111/j.1440-1746.2010.06546.x21199526 [Google Scholar] [CrossRef] [PubMed]
[3]. Glebe D, Bremer CM, The molecular virology of hepatitis B virusSemin Liver Dis 2013 33(2):103-12.Available from: https://www.thieme-connect.com/DOI/DOI?10.1055/s-0033-134571710.1055/s-0033-134571723749666 [Google Scholar] [CrossRef] [PubMed]
[4]. Datta S, Banerjee A, Chandra PK, Biswas A, Panigrahi R, Mahapatra PK, Analysis of hepatitis B virus X gene phylogeny, genetic variability and its impact on pathogenesis: implications in Eastern Indian HBV carriersVirology 2008 382(2):190-98.10.1016/j.virol.2008.09.00718952249 [Google Scholar] [CrossRef] [PubMed]
[5]. Barbini L, Tadey L, Fernandez S, Bouzas B, Campos R, Molecular characterization of hepatitis B virus X gene in chronic hepatitis B patientsVirol J 2012 9(1):13110.1186/1743-422X-9-13122769058 [Google Scholar] [CrossRef] [PubMed]
[6]. Park YM, Jang JW, Yoo SH, Kim SH, Oh IM, Park SJ, Combinations of eight key mutations in the X/preC region and genomic activity of hepatitis B virus are associated with hepatocellular carcinomaJ. Viral Hepat 2014 21(3):171-77.10.1111/jvh.1213424344773 [Google Scholar] [CrossRef] [PubMed]
[7]. Jang JW, Chun JY, Park YM, Shin SK, Yoo W, Kim SO, Mutational complex genotype of the hepatitis B virus X /precore regions as a novel predictive marker for hepatocellular carcinomaCancer Sci 2012 103(2):296-304.10.1111/j.1349-7006.2011.02170.x22136288 [Google Scholar] [CrossRef] [PubMed]
[8]. Gopalakrishnan D, Keyter M, Shenoy TK, Leena BK, Thayumanavan L, Thomas V, Hepatitis B virus subgenotype A1 predominates in liver disease patients from Kerala, IndiaWorld J Gastroenterol 2013 19(48):9294-306.10.3748/wjg.v19.i48.929424409056 [Google Scholar] [CrossRef] [PubMed]
[9]. Kaneko S, Miller RH, Feinstone SM, Unoura M, Kobayashi K, Hattori N, Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assayProc Natl Acad Sci USA 1989 86(1):312-16.10.1073/pnas.86.1.3122643103 [Google Scholar] [CrossRef] [PubMed]
[10]. Naito H, Hayashi S, Abe K, Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primersJ Clin Microbiol 2001 39(1):362-64.10.1128/JCM.39.1.362-364.200111136801 [Google Scholar] [CrossRef] [PubMed]
[11]. Kirschberg O, Schüttler C, Repp R, Schaefer S, A multiplex-PCR to identify hepatitis B virus-genotypes A-FJ Clin Virol 2004 29(1):39-43.Available from: https://www.journalofclinicalvirology.com/article/S1386-6532(03)00084-2/fulltext10.1016/S1386-6532(03)00084-2 [Google Scholar] [CrossRef]
[12]. Kyte J, Doolittle RF, A simple method for displaying the hydropathic character of a proteinJ Mol Biol 1982 157(1):105-32.10.1016/0022-2836(82)90515-0 [Google Scholar] [CrossRef]
[13]. Zhang X, Zhang H, Ye L, Effects of hepatitis B virus X protein on the development of liver cancerJ Lab Clin Med 2006 147(2):58-66.10.1016/j.lab.2005.10.00316459163 [Google Scholar] [CrossRef] [PubMed]
[14]. Bouchard MJ, Schneider RJ, The enigmatic X gene of hepatitis B virusJ Virol 2004 78(23):12725-34.10.1128/JVI.78.23.12725-12734.200415542625 [Google Scholar] [CrossRef] [PubMed]
[15]. Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in IndiaJ Med Virol 2010 82(7):1115-25.10.1002/jmv.2177420513073 [Google Scholar] [CrossRef] [PubMed]
[16]. Lee GH, Wasser S, Lim SG, Hepatitis B pregenomic RNA splicing-the products, the regulatory mechanisms and its biological significanceVirus Res 2008 136(1-2):1-7.10.1016/j.virusres.2008.05.00718579251 [Google Scholar] [CrossRef] [PubMed]
[17]. Fleischhacker M, Schmidt B, Circulating nucleic acids (CNAs) and cancer-a surveyBiochim Biophys Acta 2007 1775(1):181-232.10.1016/j.bbcan.2006.10.00117137717 [Google Scholar] [CrossRef] [PubMed]
[18]. Chattopadhyay S, Das BC, Kar P, Hepatitis B virus genotypes in chronic liver disease patients from New Delhi, IndiaWorld J Gastroenterol 2006 12(41):6702-06.10.3748/wjg.v12.i41.670217075988 [Google Scholar] [CrossRef] [PubMed]
[19]. Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK, Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinentJ Gastroenterol Hepatol 2002 17(2):165-70.10.1046/j.1440-1746.2002.02605.x11966946 [Google Scholar] [CrossRef] [PubMed]
[20]. Kidd-Ljunggren K, Oberg M, Kidd AH, The hepatitis B virus X gene: analysis of functional domain variation and gene phylogeny using multiple sequencesJ Gen Virol 1995 76(Pt 9):2119-30.10.1099/0022-1317-76-9-21197561749 [Google Scholar] [CrossRef] [PubMed]
[21]. Lin X, Xu X, Huang QL, Liu YQ, Zheng DL, Chen WN, Biological impacts of “hot-spot” mutations of hepatitis B virus X proteins are genotype B and C differentiatedWorld J Gastroenterol 2005 11(30):4703-08.10.3748/wjg.v11.i30.470316094714 [Google Scholar] [CrossRef] [PubMed]
[22]. Shinkai N, Tanaka Y, Ito K, Mukaide M, Hasegawa I, Asahina Y, Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2J Clin Microbiol 2007 45(10):3191-97.10.1128/JCM.00411-0717652471 [Google Scholar] [CrossRef] [PubMed]
[23]. Wang D, Cai H, Yu WB, Yu L, Identification of hepatitis B virus X gene variants between hepatocellular carcinoma tissues and pericarcinoma liver tissues in Eastern ChinaInt J Clin Exp Pathol 2014 7(9):5988-96. [Google Scholar]