Carcinoma Cervix is the fourth most common cancer among women with an estimated 5,28,000 new cases every year in the world [1]. Till date, out of more than 170 genotypes of HPVs, there are 40 mucosal genotypes which infect the genital tract and spread through sexual contact [2,3]. Based on their association with cervical and other anogenital cancers, mucosal HPVs are grouped as high risk and low risk genotypes. Low risk HPVs include genotypes 6, 11, 40, 42, 43, 44, 54, 61, 70, 72, 81, CP6108, probable high risk genotypes include 25, 53, 66 and, HR HPVs include genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82 [4]. HPV-16 and -18 together are responsible for about 70% cases of CaCx occurring in every region of the world [5].
Cervical HPV infection is contracted by mucosal contact during sexual intercourse and it is commonly present in young women after the onset of sexual activity. The majority of cervical HPV infections get spontaneously cleared within a few years [6]. However, the longer the duration of cervical HPV infection (persistence), the lesser are the chances that a patient can clear his/her infection. The progression from HPV infection to HPV persistence to the development of high grade lesions and ultimately cervical cancer appears to take, on an average, up to 15 years; making invasive CaCx a preventable disease [7].
Cervical Pap smear test is the routinely used screening method for identification of HSILs {Cervical Intraepithelial Neoplasia (CIN-2 and 3)} and CaCx. Since, its introduction, the Pap smear has helped reduce cervical cancer incidence and mortality rates by roughly half to two thirds [8]. However, the diagnostic confirmation is obtained on histopathological examination of cervical biopsy samples. SCC and adenocarcinoma are the two most common histological subtypes of CaCx and both are related to cervical HPV infection in more than 99% cases [9].
There are FDA approved HPV kits for detection of HR HPV in DNA isolated from cervical samples. The recently introduced kit Aptima HPV assay detects mRNA expressed by HR HPVs, thus making this test more specific than others [10]. In the cluster randomised trial, conducted by Sankaranarayanan R et al., it was concluded that in low resource settings, even a single round of HPV testing may lead to significant reduction in number of advanced cervical cancer and deaths from it [11].
Recently, an in house nested multiplex PCR has been devised to detect mucosal HPVs as a pool and simultaneously tried to type the two most common HPV genotypes-16 and -18, in CaCx [12]. In the study, lower HPV positivity (approximately 70%) in Formalin-Fixed Paraffin-Embedded (FFPE) samples of cervical SCC was attributed to formalin mediated degradation of DNA. In the present study, we have aimed at evaluating this in house nested PCR protocol for simultaneous detection of HPV, and typing of genotypes 16 and 18 in cervical scrape/biopsy samples and to correlate histological/cytological findings of these samples with the PCR findings.
Materials and Methods
This prospective cohort study was done on 63 females in the age group 21-70 years attending outpatient Department of Gynaecology, Sir Sunderlal Hospital, BHU, Uttar Pradesh, India from March 2015 to August 2016. Punch biopsy and cervical scrapes were collected from women who presented with unhealthy cervix (group A) and with apparently healthy cervix (group B).
Inclusion criteria: Married women, >20 years of age those who gave voluntary consent for the study. The women, who presented with blood stained vaginal discharge or pain abdomen and on per speculum examination revealed unhealthy cervix enrolled in group A while in group B, women who presented with complaints of vaginal discharge, irregular menses with apparently healthy looking cervix were included.
Exclusion criteria: Pregnancy, inadequate sample for Pap smear and non retrieval of DNA.
Cervical scrapings were taken with the aid of Ayre’s spatula (sterilised, disposable). Scrapings were first spread on sterile grease free slide and fixed for modified Pap staining and the remaining cells over the spatula, were washed off inside vial containing sterile Phosphate Buffer Saline (PBS) maintained at pH 7.4 for DNA based study. Cell pellet obtained after centrifugation of PBS solution were resuspended in 200 μL EDTA buffer and preserved at 4o C till further processing. Punch biopsy samples, when indicated were taken and divided into two different vials containing 10% formalin and PBS, for histological examination and DNA isolation respectively.
All the Pap smears were read by two independent observers and grading was done on the basis of Bethesda classification 2001 [13]. The formalin fixed biopsy specimens were examined and paraffin embedded blocks were prepared and further subjected to routine haematoxylin and eosin staining for light microscopic studies as per standard protocol [14].
The DNA isolation protocol was optimised using classical phenol choloform method with few modifications [15]. The nested multiplex PCR protocol standardised for simultaneous detection of HPV was performed using primers targetting 134 bp L1 capsid gene and typing of genotypes16 and 18, targeting E6/E7 gene as described previously [Table/Fig-1,2] [12,16,17]. DNA isolated from SiHa and Hela Cell line were used as positive control for HPV 16 and 18 respectively which were procured from NCCS Pune, Maharashtra, India.
Primers for first round PCR targeting consensus region of L1 and E6/ 7 (134 bp).
| S.no. | Code | Target gene | Oligo sequence | Expectedsize ofamplicons | Reference |
|---|
| 1 | GP5+ | L1 | 5’- TTT GTT ACT GTG GTA GAT ACT AC-3’ | 142 bp | de Roda Husman AM et al., [16] |
| 2 | GP6+ | 5’-GAA AAA TAA ACT GTA AAT CAT ATT C-3’ | de Roda Husman AM et al., [16] |
| 3 | GP-E6-3F | E6/7 | 5’- GGG WGK KAC TGA AAT CGG T -3’ | 630 bp | Sotlar K et al., [17] |
| 4 | GP-E7-5B | 5’-CTG AGC TGT CAR NTA ATT GCT CA-3’ | Sotlar K et al., [17] |
| 5 | GP-E7-6B | 5’-TCC TCT GAG TYG YCT AAT TGC TC-3’ | Sotlar K et al., [17] |
Primers for nested round PCR targeting consensus region of E6/7 and L1(134 bp).
| S.no. | Code | Target gene | Oligosequence | Expected amplicons | Reference |
|---|
| 1 | mGP5+ | L1 | 5’- GTT ACT GTT GTT GAT ACT AC-3’ | 134 bp | Prakash Pet al., [12] |
| 2 | mGP6+ | 5’-ATA AAC TGTAAA TCA TAT TC-3’ | Prakash Pet al., [12] |
| 3 | 16 F | E6/ 7 | 5’-CAC AGT TAT GCA CAG AGC TGC-3’ | 457 bp | Sotlar Ket al., [17] |
| 4 | 16 R | 5’-CAT ATA TTC ATG CAA TGT AGG TGT A-3’ | Sotlar K et al., [17] |
| 5 | 18 F | 5’-CAC TTC ACT GCA AGA CAT AGA-3’ | 322 bp | Sotlar Ket al., [17] |
| 6 | 18 R | 5’-GTT GTG AAA TCG TCG TTT TTC A-3’ | Sotlar Ket al., [17] |
The 142 bp L1 capsid gene product obtained after Pan HPV MY/ GP+ nested PCR using primers targeting L1 capsid gene from select SCC tissue (n=2), which did not yield HPV-16/-18 specific amplicons (n=2) [Table/Fig-3] [16,18]. The amplicons were submitted to SciGenom Labs, Cochin, Kerala, India for sequencing using Sanger’s sequencing method. The sequences obtained were first aligned and subsequently analysed with the help of online bioinformatic tool NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to know the HPV genotypes associated with the samples.
Primers for amplification of HPV L1 (142 bp) gene sequence.
| S.no. | Code | Oligosequence* | Expectedamplicons | Reference |
|---|
| 1 | MY11 | 5’-GCM CAG GGW CAT AAY AAT GG-3’ | 452 bp | Bauer HM and Manos MM., [18] |
| 2 | MY09 | 5’-CGT CCM AAR GGA WAC TGA TC-3’ | Bauer HM and Manos MM., [18] |
| 3 | GP5+ | 5’- TTT GTT ACT GTG GTA GAT ACT AC-3’ | 142 bp | de Roda Husman AM et al., [16] |
| 4 | GP6+ | 5’-GAA AAA TAA ACT GTA AAT CAT ATT C-3’ | de Roda Husman AM et al., [16] |
Statistical Analysis
The calculation of percentage was performed using Microsoft Office Excel 2007. The chi square test was applied to calculate the significance of association of HPV 16/18 positivity with different pre invasive lesions of CaCx. A p-value less than 0.05 were considered significant.
Results
The present study is based on observations made on 49 cervical smears and 14 cervical biopsies obtained from 63 subjects, age 21-70 years enrolled for the study. On physical examination cervix was found unhealthy in 47 patients (group A) while in 16 subjects it was apparently healthy (group B).
Of the 14 cases, in which cervical biopsies were studied, 13 were diagnosed as SCC while one was categorised as CIN2. Cytological diagnosis of 49 cervical scrapes has been shown in [Table/Fig-4].
Cytological diagnosis of cases subjected for Pap smear examination (n=49).
| Cytological diagnosis | Group A (n=33)n (%) | Group B (n=16)n (%) | Totaln (%) |
|---|
| NILM | 23 (69.6%) | 14 (87.5%) | 37 (75.5%) |
| ASCUS | 1 (3.0%) | 1 (6.2%) | 2 (4.1%) |
| LSIL | 6 (18.2%) | 1 (6.2%) | 7 (14.2%) |
| HSIL | 3 (9.0%) | 0 (0%) | 3 (6.1%) |
NILM: Negative for intraepithelial lesion or malignancy; ASCUS: Atypical squamous cells of undetermined significance; LSIL: Low grade squamous intraepithelial lesion; HSIL: High grade squamous intraepithelial lesion
On subjecting 13 samples of SCC of uterine cervix to nested multiplex PCR, for simultaneous detection of HPV as well as typing genotype -16 and -18, it was observed that 12 samples yielded 134 bp L1 capsid amplicons. Of these, two samples showed positivity for both HPV-16 and HPV-18. Seven CaCx cases were exclusively positive for HPV-16 whereas one was positive for HPV-18 and two were negative for both HPV-16 and HPV-18. One biopsy sample did not yield 134 bp L1 capsid amplicons however exclusively yielded 457 bp HPV-16 specific amplicons [Table/Fig-5,6].
Correlation of HPV positivity with histological diagnosis (n=14).
| Diagnosis | No. ofsamples | NMPCRE6/7 and GP+/mGP+n (%) |
|---|
| L1 capsid gene(134bp) | E6/7gene(457bp HPV-16) | E6/7 gene(322 bp HPV-18) |
|---|
| CIN1 | 0 | 0 (0%) | 0 (0%) | 0 (0%) |
| CIN2/3 | 1 | 1 (100%) | 1 (100%) | 0 (0%) |
| SCC | 13 | 12* (92.3%) | 10† (76.9%) | 3‡ (23.1%) |
| ADC | 0 | 0 (0%) | 0 (0%) | 0 (0%) |
*Two samples which were not typed as HPV-16 or 18 by nested multiplex PCR were HPV-16 and 73 on sequencing
†One HPV-16 infected CaCx sample was negative for 134 bp L1 amplicons.
‡Two HPV-18 infected CaCx tissues were also observed to be coinfected with HPV-16
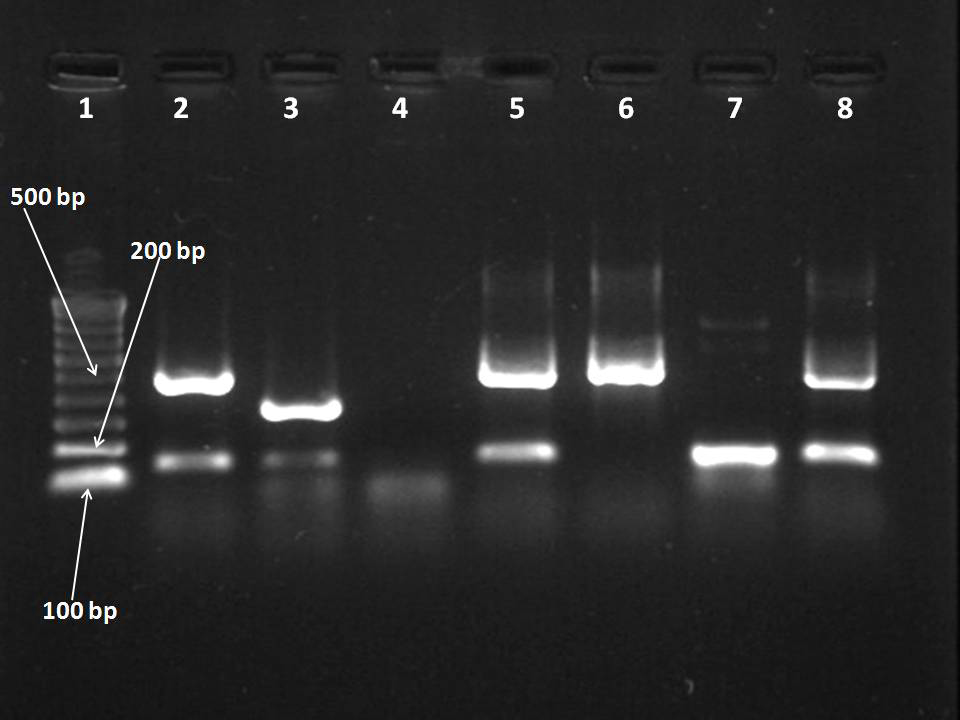
Gel picture of nested multiplex PCR for simultaneous detection of HPV and genotype -16 and -18 in biopsy samples.
Lane 1:100 bp DNA ladder
Lane 2: SiHa cell line (134 bp and457 bp)
Lane 3: HeLa cell line (134 bp and 322bp)
Lane 4: Negative control (mili Q water)
Lane 5 and 8: Positive for HPVL1 (134 bp), HPV-16(457bp)
Lane 6: Positive for HPV-16 (457bp) only
Lane 7: Positive for HPVL1 (134 bp) only
Two biopsy samples, namely Cx 14 and 30, which did not yield HPV-16/-18 specific amplicons; however, positive for 134 bp HPV L1 specific amplicons were subjected to alternative nested PCR protocol for detection of 142 bp L1 gene sequence employing MY/GP+ primers. Nested PCR product of these samples was submitted for DNA sequencing.
After correcting and aligning sequence information obtained from forward and reverse strand from amplicon of one of the sample (Cx 14), while searching for highly similar sequences using NCBI BLAST tool, the 142 bp sequence showed 100% similarity to HPV-16 sequence (accession number: JN617890.1) present in the database [Table/Fig-7].
Forward sequences obtained after MY/GP+ nested PCR from select samples.
| Sampleno | Lab code | Sequence obtained from MY/GP+ nested PCR products 6624 bp-6765 bp (HPV 16/18) | Sequencesimilarity/L1 Variant |
|---|
| 1 | Cx 14 | TTTGTTACTGTGGTAGATACTACACGCAGTACAAATATGTCATTATGTGCTGCCATATCTACTTCAGAAACTACATATAAAAATACTAACTTTAAGGAGTACCTACGACATGGGGAGGAATATGATTTACAGTTTATTTTTC | 100% withHPV 16SequenceID: gb|JN617890.1| |
| 2 | Cx 30 | TTTGTTACTGTGGTAGATACTACTAGAAGCACTAATTTTAATGTATCTGTAGGTACACAGGCTAGTAGCTCTAATTTTACGGACGTTTAAGACATGCAGAAGAATATGATTTACATTTTAATTTTC | 76% withHPV 73SequenceID: gb|KU050128.1| |
However, the sequence information obtained from forward and reverse strand from amplicon obtained from the sample Cx 30, yielded a sequence which did not showed significant similarity to sequences already submitted in the database, while searching for highly similar sequences. When nucleotide BLAST was performed searching for either more dissimilar sequences (discontinuous megablast) or somewhat similar sequences (blastn), the sequence was observed to be 76% similar to HR HPV-73 (Accession number: KU050128.1) [Table/Fig-7]. Thus the result of sequencing of PCR products revealed that one of these samples was typed as HPV-16 and whereas the other one was HPV-73.
[Table/Fig-8] presents correlation of HPV positivity with cytological diagnosis. Out of 37 cases reported as negative for NILM, 9 (24.32%) cases were positive for HPV L1 (134bp), 5 (13.5%) were positive for HPV-16. Out of two cases of ASCUS, both (100%) cases were positive for HPV L1 (134bp) and 1 (50%) case was simultaneously positive for HPV-16. Out of the seven cases presenting with LSIL, 4 (57.1%) were positive for HPV L1 and 1 (14.3%) was simultaneously positive for HPV-16. While out of three cases of HSIL, 2 (66.7%) were HPV L1 positive, 1 was positive for HPV-16 and 1 case was positive for HPV-16 as well as 18 however did not yield HPV L1 (134-bp) amplicon.
Correlation of HPV positivity with cytological diagnosis (n=49).
| Diagnosis | No. ofsamples | NMPCRE6/7 and GP+/mGP+n (%) |
|---|
| L1 capsid gene(134bp) | E6/7gene(457bp HPV-16) | E6/7 gene(322 bp HPV-18) |
|---|
| NILM | 37 | 9 (24.3%) | 5 (13.5%) | 0 (0%) |
| ASCUS | 2 | 2 (100%) | 1 (50.0%) | 0 (0%) |
| LSIL | 7 | 4 (57.1%) | 1 (14.3%) | 0 (0%) |
| HSIL | 3 | 2 (66.7%) | 2 (66.7%) | 1 (33.3%) |
On statistical analysis, the HPV detection rates as well as HPV-16/18 positivity rate in both the groups were observed to be comparable [Table/Fig-9].
Status of HPV positivity in cases subjected for cervical scrapes (n=49).
| HPVStatus | Group A (n=33b)n (%) | Group B (n=16d)n (%) | p-value |
|---|
| HPV | 13a (39.4%) | 4c (25.0%) | 0.32 |
| HPV-16 | 8e (24.2%) | 2f (12.5%) | 0.24 |
| HPV-18 | 1g (3.0%) | 0h (0%) |
χ2 test: a/b vs c/d, p>0.05; e+g/b vs f+h/d, p>0.05; Significant at the 0.05 probability level
Discussion
HPVs are most common sexually transmitted agent. Further, HR HPV infection is associated with the development of cervical neoplasia and is necessary, however not sufficient factor in the aetiology of cervical carcinoma [9,19]. Viral persistence is required for neoplastic progression, and increased risk has been associated with early high viral loads [20-22]. However, it has been observed that HR HPVs of all types are able to induce malignant tumors even when they are present at low levels [23].
HPV infection is extremely common in young women in their first decade of sexual activity. The longer the HPV infection persists, the less likely a patient can clear her infection. It has been observed that less than 10% of new infections became persistent, develops into dysplasia which may progress to precancerous lesions, usually in 5-10 years [24]. Secondly, cervical cancer most often develops in women after age 40 and is most frequent among women in their fifties and sixties [25]. In the majority, it usually regresses or does not progress, particularly among women under age 35. One study estimates that the risk of progression from moderate to severe precancerous lesions is 32% within 10 years, thus provides opportunity to screen the population at risk [26].
In India, studies on HPV testing have largely focused on detection for screening purposes [27-29]. Menon et al., reported HPV 16/18 in 76.4% of 72 cervical malignancy cases, less than half of 24 cervical intraepithelial neoplasia cases and in none of four normal cervical tissues tested [30]. Munirajan et al., detected HPV in 70% of 43 cases of CaCx of stage IIB/IIIB, with HPV-16 in 23 cases (53%), HPV-18 in four cases (13.3%), and 1 case each of HPV-33, 35 and 58 [31]. In a hospital based, case control study from Chennai, Tamil Nadu, India HPV was detected by PCR in all however one of the 205 invasive cervical carcinoma cases and in 27.7% of the 213 age matched controls. 23 different HPV types were reported, with HPV-16 being the most common type, followed by types 18 and 33 [32].
A study conducted in Andhra Pradesh, India also indicated HPV-16 and 18 to be most common HR HPV among cancer tissue, however HPV-52 was most common HR HPV observed in cervical samples of women in Medchal community indicating variation in epidemiological distribution of the virus [33]. In another study from western Uttar Pradesh and Delhi region, revealed that of the 90 cervical tumor samples screened with L1 consensus primers, 65 (72.2%) were positive for HPV, and further genotyping with type-specific primers revealed 92.3% (n=60) positivity for HPV-16 and 7.7% (n=5) positivity for HPV-18 [34].
In a study, conducted on cervical samples collected from TMH, Mumbai, India by Travasso CM et al., attempted to use novel pyrosequencing method to genotype the HPV and observed that among the cervical cancer samples, 96.9% showed the presence of HR HPVs, including HPV-16 (73.8%), HPV-18 (10.77%), HPV-33 (3.07%), HPV-31 (1.53%) and HPV-45 (1.53%) [35]. In a study from Guwahati, Assam, in which HPV detection was attempted in 107 cervical cancer patient samples using nested multiplex PCR assays for detection of 13 HR HPV and five low risk HPV types, HPV was confirmed in 105 samples. The presence of six ‘carcinogenic’ HPV types, HPV-16 (88%), -18 (15%), -31(4%),-45 (3%), -59 (4%), -58(1%), and one non carcinogenic, HPV-6/11 (6%), was recorded [36].
While observing the HPV genotype distribution in cervical intraepithelial neoplasia among HIV-infected women in Pune, Maharashtra, India the carcinogenic HPV genotypes in declining order of prevalence were HPV-16,-56,-18,-39,-35,-51,-31,-59,-33,-58,-68,-45 and -52 [37]. In a similar study, in which HPV genotype distribution of HPV were observed in HIV positive females, HPV-16 was the most common type, detected in 42% of HPV positive women, followed by HPV-45 (15%), HPV-18/52/31/58 (11.5% each), and HPV-33 (7.6%). The corresponding figures in the control group were as follows: HPV-16 (66.6%), HPV-45/-18/-31 (16.6% each), and HPV-33/-58/-68 (8.3% each) [38]. HPV-73 has also been documented in a recent study from AIIMS, New Delhi, India. Thus, till date, all 15 HR HPV genotypes have been reported from various centres in India [39].
There are various in house PCR based protocols, which are used to detect HPVs in cervical scrapes and biopsy tissue samples. Among these, HPV detection is usually done by PCR employing General Primer (GP5+/ GP6+) targeting consensus sequence of conserved region of L1 capsid gene present in different genotypes of HPV [16]. Sotlar K et al., was first to put forth the concept multiplexing the primers for nested PCR based genotyping all the important HPV types in clinical samples by targeting E6/7 gene of the virus. The use of nested multiplex PCR protocol using MY09-MY11, mGP5+-mGP6+ and other primers targeting the E6/E7 region, has been shown to increase the overall sensitivity compared to that of each primer pair alone [17].
In this study, we first attempted to correlate histological findings of 14 cervical biopsies with HPV positivity and association of genotype 16 and 18. Of these 14 cases, all 13 cases of SCC cervix were observed to be HPV positive and of these 12 were HPV16/18 positive and one was positive for HPV-73. Therefore, all the cases of SCC cervix tissues yielded HPV specific amplicons with the nested multiplex PCR protocol. As 80-90% of carcinoma of uterine cervix is histologically documented as SCC and it is this histological types which is almost 100% associated with HR HPVs [40].
Further, after multiplexing the two different target genes for detection and typing of HPV i.e. L1 and E6, we were able to detect HPV in all samples of SCC of uterine cervix. In the study, there were two CaCx samples which yielded either of the two target sequence i.e., 134 bp L1 or 457 bp HPV16 specific E6/7 gene product. As this variant of PCR can be standardised in house and could prove to be a cost effective protocol.
Upon attempting to observe HPV positivity in cervical scrapes samples of females collected in PBS, it was observed that HPV 16/ 18 positivity among both the study group was comparable indicating importance of DNA based screening among women irrespective of physical appearance of cervix.
It has been observed that by targeting two different genes of HPV namely L1 and E6/7 helped in detection of those HPV associated cases which did not yield 134 bp consensus sequence amplicons both in cervical scrape and biopsy samples. However, as the present PCR protocol does not type the 13 less common high risk genotypes of HPV, one has to rely on MY/GP+ nested PCR and sequencing to elucidate the presence of other possible high risk genotype in the sample.
Limitation
In this study, we were not able of follow-up the subjects for observing the persistence of HPV infection and or progression of lesions.
Conclusion
The PCR protocol used in this study for simultaneous detection of HPV and typing of two most common genotypes HPV-16 and HPV-18 appears to be an appropriate HPV detection tool in cervical samples. The results indicates that the PCR protocol may be further evaluated for its utility in screening of CaCx, in a prospective multicentric study with follow-up, on large number of subjects.
Ethical approval: The study was approved by the Institute Ethics Committee of Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India vide letter Dean/2014-15/1112 dated 20-03-2015.
Authors’ Contributions
PP and AGK conceived of the study. NG and NRA performed the sampling and data collection. NG, PP and AGK participated in the analysis of the samples, and data management. NG and PP drafted the manuscript. All of the authors read the manuscript and approved the final version prior to submission.
NILM: Negative for intraepithelial lesion or malignancy; ASCUS: Atypical squamous cells of undetermined significance; LSIL: Low grade squamous intraepithelial lesion; HSIL: High grade squamous intraepithelial lesion
*Two samples which were not typed as HPV-16 or 18 by nested multiplex PCR were HPV-16 and 73 on sequencing
†One HPV-16 infected CaCx sample was negative for 134 bp L1 amplicons.
‡Two HPV-18 infected CaCx tissues were also observed to be coinfected with HPV-16
χ2 test: a/b vs c/d, p>0.05; e+g/b vs f+h/d, p>0.05; Significant at the 0.05 probability level