Kidney stones are classified into metabolic and infectious stones and are extremely frustrating common health complications in people around the world. Rather expanding much as 1 out of 10 individual is affected by the urinary stone disease but the mechanism behind the formation of stones is not clearly understood [1]. Stone formation causalities are not just limited to supersaturation (beyond the dissolution limit) of CaOx or Calcium-Phosphate (CaPhos) components but that can be multifactorial phenomenon is now easily perceived. Bacteria can be one of the responsible factors in stone generation that may not be denied. Attention towards the bacteria for different prospective like raising the confirmation of intestinal bacterium Oxalobacter formigenes had been negatively associated with calcification and recurrence of stones [2].
Previously, the urinary tract was considered to be sterile; however, urine culturing protocols and Next Generation Sequencing (NGS) techniques proved the presence of microbes in urine [3]. Knowledge about the bacterial invasion and/or localisation inside the CaOx stones is very scanty. The exploration of bacterial imprints inside the kidney stones has been getting the attention most of the researchers across the globe and methodologies mostly acquired, are direct detection methods such as cultivation and microscopy. The present study aimed at characterising the chemical composition of kidney stones and assesses total bacterial diversity in these stones using 16S rRNA gene clone library approach. The present study involved in the identification of bacterial composition of kidney stone and also in understanding their putative metabolic activities involved in pathophysiology formation of kidney stone.
Materials and Methods
Study Subject Recruitment and Sample Collection
In the present retrospective study, subjects recruitment and sample collection were done during November 2012 to November 2015. A total of 24 symptomatic recurrent kidney stone male patients were recruited and 24 hour urine, stool, surgically removed kidney stones and signed informed consent was collected as per with Institutional Ethical Committee, National Centre for Cell Science (IEC-NCCS), Maharashtra, India. Scrutiny, recruitments, inclusion and exclusion criteria and consent for present study as a part of the case-control study [4,5]. Briefly, we recruited the hospitalised, and under observation kidney stone disease subjects who had been confirmed their status by ultra-imagine (like either by X-ray, KUB, Ultrasound, and Tomography etc.,). Male subjects (22-51-year-old) with multiple stone episodes were included. No any gastrointestinal disorder and/or surgery, usage of antibiotics or probiotic preparations for last three months before enrolment were set for exclusion criteria. The 24 hour urine and stool samples were utilised for oxalate quantification and bacteriological diversity analysis, respectively; while the stones were used here for further analysis [4,5].
Chemical Analysis of Kidney Stones
A total of 120 surgically removed kidney stones (S1_p1 and S1_p2) were collected in sterile containers; surface washed with 70% ethanol, later with sterile distilled water and consequently air dried. Each kidney stone was aseptically pulverised into powdered form, part of which was analysed by FTIR spectroscopy without KBr method to identify the stone type [6]. Briefly, each part of stone was placed in the Infra Red (IR) beam of the Bruker FTIR ATR Tensor 37 spectrometer (Bruker Inc, USA) and spectra were obtained in transmittance mode from 4000 to 400 cm-1; 32 scans were averaged with a 4 cm-1 resolution for each spectrum. The resulting spectra obtained for all 120 stones were compared to the spectrum of standard CaOx (Sigma-Aldrich Co., USA).
Bacterial Diversity Analysis
Stone powder of all stones derived from the single patient was pooled and used for genomic DNA isolation as previously described, which was further subjected to 16S rRNA gene clone library preparation [7]. Briefly, 16S rRNA gene was amplified using eubacteria specific primers: 27F (5′-CCAGAGTTTGATCMTGGCTCAG-3′) and 1490R (5′-GGTTACCTTGTTACGACTT-3′) [8]. Following conditions were utilised for PCR amplification: Initial denaturation; 94°C for 5 minutes followed by 28 cycles of denaturation 94°C for 1 minute, annealing; 55°C for 0.5 minutes and extension 72°C for 1 minute, and a final extension; 72°C for 7 minute. The resulting PCR products were cloned, sequenced, and good quality clone sequences were obtained [9]. Briefly, the 16S rRNA gene products were purified with Gene Elute Gel Extraction Kit (Sigma-Aldrich, St Louis USA), ligated into pCR4® TOPO vector supplied with the TOPO TA cloning kit (Invitrogen, San Diego, USA) and transformed into One Shot TOPO10 electro-competent Escherichia coli (E. coli) (Invitrogen, San Diego, USA) following the manufacturer’s instructions. Sterile Luria Bertani agar with 50 μg/mL (W/V) of kanamycin (Sigma-Aldrich Co., USA) was used for selection of the transformed cells, which were incubated for 16 hours at 37°C M13F (5’-GTAAAACGACGGCCAGT- 3’) and M13R (5’-GCGGATAACAATTTCACACAGG-3’) primers were used for screening and sequencing of these clones as per the manufacturer instructions (Invitrogen, San Diego, USA). The bidirectional sequencing was performed on ABI 3730XL DNA analyser (Applied Biosystems Inc, USA) using the ABI Bigdye terminator version 3.1 sequencing kit as per the manufacturer’s instructions. The forward and reverse sequence reads were assembled and contigs were obtained using ChromasPro software (www.technelysium.com.au/ChromasPro.html). All sequences were checked for chimeric artifacts by Mallard program, using E. coli U000096 as a reference sequence [10]. All of these good quality sequences are deposited to National Center for Biotechnology Information (NCBI) GeneBank under the accession numbers KM581286-KM581327 and Basic Local Alignment Search Tool (BLAST) analysed using curated type strain 16S rRNA gene database of EzTaxon-e server (http://www.ezbiocloud.net) [11]. Results were recorded for the taxonomy of each clone sequences with the similarity of 97% cut off in BLAST analysis. Further, species level identification of each clone sequences based on the highest blast analysis similarity value was used to point out the diversity in kidney stones.
Results
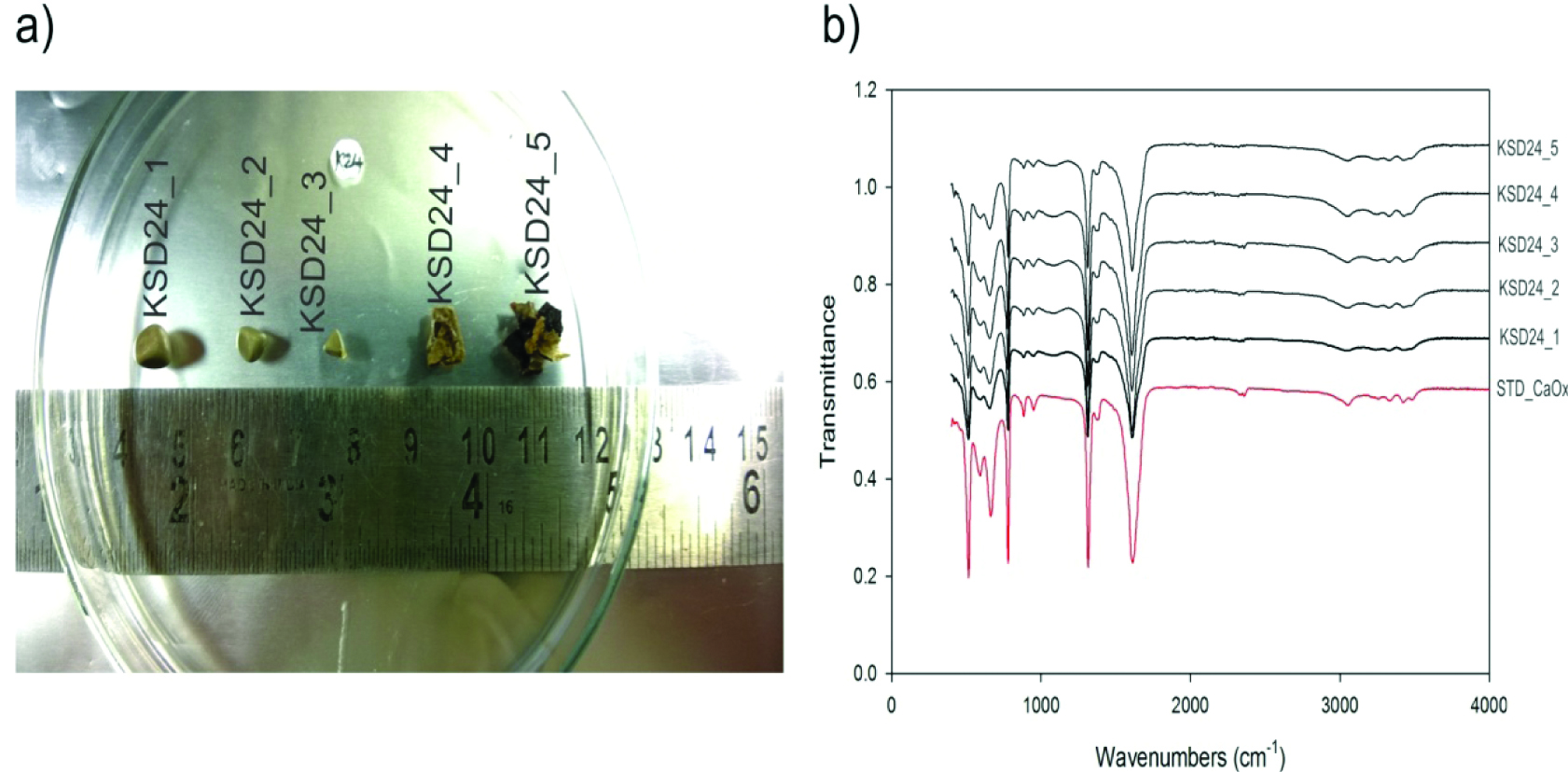
All stones analysed were found to be pure CaOx stones as revealed by FTIR analysis [Table/Fig-1]. Wavenumber and transmittance of all the analysed stones are given in supplementary excel sheet (S2). The study subjects were found to be carrying only metabolic kidney stones of CaOx origin. As per the basic assumption, the involvement of bacteria and their growth components may induce the CaOx stone formation. In connection with this, if there are any bacteria present inside the CaOx stone has to be identified at the very first place. So, analysis and taxonomic characterisation of bacteria using 16S rRNA gene clone library preparation is now a gold standard approach to study the total bacterial diversity of a variety of samples [12].
Surgically removed kidney stones and their analysis. Representative stones from subject_KSD24: a) Photograph of kidney stones from subject_KSD24; b) FTIR spectra of respective stones.
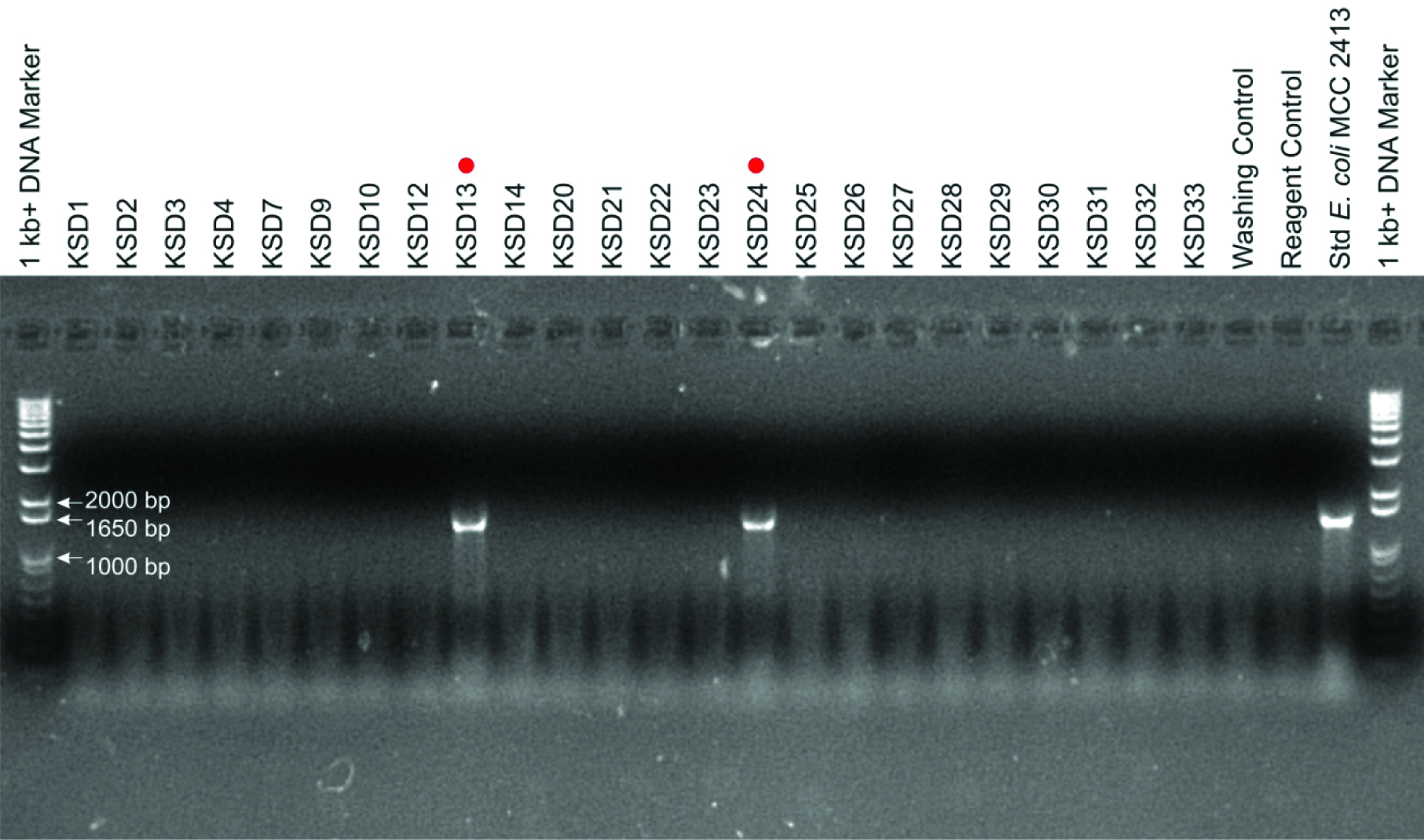
Following the drench of clues in literature here adverted the molecular diagnosis method for microbial presence inside the kidney stone nidus using 16S rRNA clone library preparation [Table/Fig-2]. Out of 24 pooled kidney stones tested, two were positive for 16S rRNA gene amplification, consequently, we generated two 16S rRNA gene clone libraries [Table/Fig-3]. By analysing 42 clone sequences we report the presence of bacteria in kidney stones, most of which were belonged to the genus Bacillus [Table/Fig-4]. In addition, bacteria belonging to genera Acinetobacter, Enterococcus, Leucobacter, Prolinoborus and Streptococcus and respective species were also observed in varying distribution in these stones.
Major steps of the methodology followed for the molecular imprint analysis of kidney stone nidus for present study.
Agarose gel fingerprint of bacterial imprints through 16S rRNA gene amplification as a molecular marker for present study. Total 02 positive samples (KSD13 and KSD24) for amplification out of 24 kidney stones, and size of amplicon products around 1500 bp which is correspondence with the 1 Kb+ DNA marker/ladder are as displayed in 1% agarose gel after electrophoresis.
Identification of 16S rRNA gene library clones, their putative products as a risk factor for kidney stone formation and oxalic acid metabolism abilities.
| S. No. | Clone Nearest Hit | No. of Clones | Putative metabolite or activity as potential risk factor† | Oxalic acid metabolism ability† |
|---|
| 1 | Acinetobacter lwoffii NCTC 5866 | 2 | Acid tolerance | Utilisation |
| 2 | Bacillus anthracis Ames | 5 | Surface cell wall protein, Spore coat | Utilisation |
| 3 | Bacillus anthracis ATCC® 14578™ | 20 | Surface cell wall protein, Spore coat | Utilisation |
| 4 | Bacillus cereus ATCC®14579™ | 1 | Surface cell wall protein, Spore coat | Utilisation |
| 5 | Enterococcus faecium ATCC® 19434™ | 2 | Acid tolerance, Exopolysaccharides | Degradation |
| 6 | Leucobacter chromiireducens subsp. chromiireducens L-1 | 4 | Acid tolerance, Exopolysaccharides | Tolerance and Utilisation |
| 7 | Leucobacter kyeonggiensis F3-P9 | 1 | Acid tolerance, Exopolysaccharides | Tolerance and Utilisation |
| 8 | Prolinoborus fasciculus CIP 103579T | 6 | Spore coat, Biodegradation of proline | NA |
| 9 | Streptococcus anginosus CCUG 39159 | 1 | Biofilm formation, Calcium ion aggregation | Degradation |
†Based on literature survey, NA-Not Any report found
Discussion
In the present study, 16S rRNA gene (as a molecular marker) was used for the identification and estimating the diversity of eubacteria inside the CaOx containing metabolic stones. Use of clone library generation approach suggests that the molecular profiling of any biological entities can be possible and may give hopes from abnormally produced clinical specimens even in physically hard in nature. Recently, these technique has also been adopted to characterise the bacterial diversity associated with gall stone; hence, we have adopted this approach to characterise CaOx stone associated bacterial diversity [13,14].
The bacteria present in nidus of kidney stone possess one or more abilities which may make them a potential contributor to the development of recurrent oxalate kidney stone [Table/Fig-4]. The present results are strengthening by another in-vitro study, in which role of Gram-positive and Gram-negative bacteria in lithiasis of CaOx stones has been dealt in mechanical and illustrative ways [15]. The genus Bacillus includes many spore-forming species; bacterial endospores can be a reason to initiate the deposition of calcium carbonate [15]. To the best of our knowledge, this is first reported to show the presence of bacteria other than Escherichia, Proteus, Staphylococcus, Providencia, and Klebsiella which are commonly associated with the nidus of stone. Surprisingly, we did not detect any routine Enterobacteriaceae members such as E. coli in all stones studied. The present study observation indicates that indeed there could be many more bacteria associated (such as those listed in with the development of metabolic stone especially in case of recurrent episodes of stones [Table/Fig-4]. However, further studies on a large number of samples are required to show whether members of Enterobacteriaceae are really absent in all the cases or they are present along with bacteria that we were able to detect in the stone.
Sometimes stones formation is exclusively associated with bacteria as in the case with infectious stones and is considered to be a consequence of a Urinary Tract Infection (UTI) [16]. Presence of bacteria inside the nidus of kidney stones has already been demonstrated but the exact role of these bacteria in the nucleation and growth of stone is poorly understood [17]. However, some explanations suggest that colonisation of bacterial cells inside the nidus, would exhibit either renal tubular acid tolerance, degradation of amino acids or changing the pH of microenvironment milieu by ammonia production through urease activity [18-20]. Another possible explanation towards the role of bacteria in the formation of stone is their ability to inhibit and/or alter the inhibitors of kidney stone formation [21].
Presence of bacteria in stones provides the perception that in hyperoxaluric condition, bacteria are not only found in the kidney but they may participate in the formation of kidney stones and other urological diseases [22]. Growing evidence support the fact that either microbial cells or their metabolic products are associated with the development of metabolic as well as infectious kidney stones. In the best of our knowledge, this is the first study to report the presence of bacteria in surgically removed metabolic stones. Thus, studies such as the present study are helpful to find out the exact bacterial composition of stone and are essential to predict the stone type as hypothesised earlier. Furthermore, the study suggests the need of combinatorial approach may be in the form of antibacterial therapy along with surgical procedures to provide an exact remedy for recurrence of stones.
Limitation
Sample size and gender bias where the intrinsic properties of present study that limits the statistical power calculations. Such molecular detection methods are based on polymerisation reactions and some of the extracted molecules (e.g., mineral components) in extracted DNA from a kidney stone may acts as inhibitor for these PCR. More sample size and different procedures in PCR activity will definitely overrule in further pilot experiments.
Conclusion
Here the study reports the presence of bacteria and its diversity in CaOx stones that were previously unfamiliar which may help urologist in understanding the possible association of bacteria with CaOx stones. These bacteria may be responsible for recurrent episodes of stones and hence, there is a need of suitable antibacterial therapy along with surgical procedures for curing the kidney stone disease.
†Based on literature survey, NA-Not Any report found