Fungal corneal infection or “mycotic keratitis” is a major cause of preventable blindness especially in tropical and sub tropical countries. In India, about 60,000 cases of mycotic keratitis and about 10,000 resultant corneal transplants occur each year [1,2]. The reported incidence of mycotic keratitis is 17-36% worldwide, whereas, it is about 44-47% in India [3-6]. Corneal trauma with plant or animal materials or dust particles due to either agricultural activity or animal husbandry, blocked nasolacrimal duct, poor hygienic practice of soft contact lens are the most common predisposing factors of keratomycosis [7,8]. Majority of the patients come from rural areas [7]. Fungal keratitis or keratomycosis, without early and prompt treatment, may lead to either loss of vision or severe visual impairment [2,3]. In contrast to bacterial keratitis, signs and symptoms of mycotic keratitis are mild and moderate in the early stage due to mild degree of inflammation; but, later on it leads to suppuration and ulceration [1].
Invasive fungal infections cause severe illness in the patients. IFIs are predominantly seen in immunocompromised individuals, but several cases were also reported in immunocompetent individuals [9,10]. Patients with IFIs show signs and symptoms of febrile illness that continue even after prolonged broad spectrum antibiotics treatment and lesions are visible in radiology report [9]. Systemic predisposing factors like diabetes, Human Immunodeficiency Virus (HIV) infection, cancer and treatment for a bacterial infection with antibiotics or steroids influence the occurrence of IFIs [10]. As the number of reported cases of IFIs are few and data is scanty, exact prevalence from India is hard to define. Early and accurate diagnosis with proper treatment is essential for preventing morbidity and mortality in these patients.
Fungal species make up for nearly 7% (611,000 species) of all eukaryotic species on earth and are widely distributed in plant debris, soil, dust and other organic substrates among which about 600 species are human pathogens [11,12]. Although, more than 105 species of fungi are known to cause eye infections, majority are caused by members of Aspergillus spp. and Fusarium spp. across India and South East Asia, among which, Aspergillus spp. was reported as the most predominate and prevalent one in Northern India [13,14]. The other commonly isolated filamentous fungi from fungal corneal infections are Alternaria spp., Penicillium spp. and Curvularia spp. [1]. Opportunistic yeasts like Candida specially C. tropicalis and C. albicans or filamentous fungi like Aspergillus spp. are the most predominant genera of fungus involved in IFIs in India and worldwide [15,16]. Other fungi such as Fusarium, Trichosporon and Malassezia spp., which were previously considered as non-pathogenic for humans or causing diseases sporadically are now considered as leading fungal pathogens for IFIs [15,16].
Keratomycosis and IFIs can be misdiagnosed leading to severe complications. In the early stage when symptoms are mild; accurate and rapid diagnosis is the key feature for the management of fungal keratitis and invasive infections. Previously, the laboratory diagnosis of fungal keratitis and IFIs were only made by direct microscopy of corneal scraping, tissues or body fluids with Potassium Hydroxide (KOH) mount and isolation in culture [17]. As the sensitivity of direct microscopy is low, the staining with calcofluor white and blankophor increases the sensitivity to some extent [18]. Culture remains the gold standard for diagnosis of fungal keratitis and IFIs [19]. In some cases, fungal culture isolates may not be identified accurately on the basis of morphology and microscopy. In case of keratomycosis or infections by highly invasive fungi like Pythium spp., the fungus can be misidentified as contaminant or Aspergillus spp. leading to incorrect therapy and severe morbidity, sometimes with fatal consequences. In those cases, molecular characterisation of isolates become more helpful in identification, which is essential for providing appropriate antifungal therapy and avoiding bad prognosis [20].
The present study aims to identify the fungi collected from the patients of fungal keratitis and IFIs (diagnosed by conventional methods) using Polymerase Chain Reaction (PCR) assay for both ITS regions of ribosomal RNA, nucleotide sequencing, analysis using National Center for Biotechnology Information-Basic Local Alignment Search Tool (NCBI-BLAST) against available data base in gene bank and to compare the results with the results of conventional methods of identification (direct microscopy and phenotypic identification from culture isolates).
Materials and Methods
Fungal Culture Isolates
A prospective study conducted over a period of one year (March 2016 to March 2017) was carried out at the Department of Microbiology, All India Institute of Medical Science (AIIMS), Delhi, India. The fungal culture isolates on Sabouraud’s Dextrose Agar (SDA) (only filamentous fungi) from corneal scrapings of 24 keratomycosis patients (diagnosed by conventional methods) attending the OPD/ causality services of Dr. Rajendra Prasad Centre for Ophthalmic Sciences, AIIMS. Fungal isolates (only filamentous fungi) from seven IFI patients (diagnosed by conventional methods) admitted to different wards of the AIIMS hospital were included in this study. Patients of clinically suspected fungal keratitis and invasive infections by filamentous fungi were included in this study. Patients of either Acanthamoeba keratitis, bacterial keratitis or viral keratitis and IFI patients with yeast infections were excluded from this pilot study. The age of the patients of keratomycosis ranged from 6-79 years with the mean age of 39 years with predominance of male patients were 22 (92%). The age of the patients of IFIs varied from 24-52 years with the mean age of 39 years out of which 3 (43%) were males and 4 (57%) were females. All procedures performed in this study were in accordance with the ethical clearance of the institute. The fungal culture isolates were sub-cultured on SDA (HiMedia, India) with antibiotic gentamicin (0.02 mg/mL). Two tubes were taken per isolate, among which one was incubated at 25°C and other at 37°C, till the growth appeared [21,22].
Macroscopic and Microscopic Examination of the Culture
The culture slants were examined for morphological features, color, texture, diffusible pigments and spore features etc. Lactophenol Cotton Blue (LPCB) mount was prepared and visualised under a light microscope with 10X and 40X objectives for microscopic examination [23].
Molecular Methods
DNA isolation from fungal culture isolates: DNA was isolated from the 31 fungal culture isolates using the protocol described by Lee SB et al., with slight modifications [24]. Briefly, primary fungal culture isolates obtained from the patients were further subcultured on SDA plates, and kept in an incubator at 25°C and 37°C for at least 7-10 days until a full grown fungal mat was seen over the agar surface. Mycelial mat of approximately 0.3-0.5 gm was taken in a sterile mortar; liquid nitrogen was added and grounded quickly to prepare fine powder using a pestle. Fungal powder of about 0.2-0.3 gm was transferred to sterile 1.5 mL microcentrifuge tube and 600 μL of ATL lysis buffer (QIAGEN, Germany) was added. Microcentrifuge tubes were vortexed briefly and incubated at 65°C for three hours. Lysis buffer AL (200 μL) (QIAGEN, Germany) and 20 μL of proteinase K was added to the tubes, vortexed and incubated at 56°C for one hour followed by at 70°C for 10 minutes. About 4 μL of RNase A (10 mg/mL) (QIAGEN, Germany) was added to the tubes; incubated at 37°C for one hour to digest the contaminating RNA. Lysed solution was mixed with 200 μL of 100% ethanol and incubated for two minutes at room temperature. The solution was transferred to QIAamp mini-spin column (QIAGEN, Germany) and centrifuged at 8000 rpm for one minute. Tubes were washed with 500 μL of washing buffer AW1 followed by AW2 with centrifugation at 8000 rpm for one minute each. DNA was eluted with 100 μL of elution buffer AE (QIAGEN, Germany) and stored at -20°C for PCR assay.
PCR assay for molecular diagnosis: PCR assay for amplification of the genus specific Internal Transcribed Spacer (ITS) regions ITS1 and ITS2 of ribosomal DNA (~280 bp) was standardised with published primers {ITS1 region: FP: (5’-TCC GTAGGTGAACCTGCGG-3’) that hybridises at the end of 18S rDNA and RP: ITS86 (5’-GTTCAAAGATTCGATGATTCAC-3’) hybridises with the 5.8S rDNA region} and {ITS2 region: FP: ITS86 (5’-GTGAATCATCGAATCTTTGAA C-3’) that hybridises with the 5.8S rDNA region and RP: ITS4: (5’-TCCTCCGCTTATTGATATGC-3’) which hybridizes at the beginning of 28S rDNA} by changing the concentration of MgCl2 and annealing temperature until specific bands were visualised in 1.5% agarose gel [25-27]. Briefly, the protocol standardised for the PCR assay was as follows: a reaction mixture of 20 μL was prepared with 1X reaction buffer (Fermentas), 2.5 mm MgCl2, 200 μm Deoxyribonucleotide Triphosphates (dNTPs) (Fermentas), 0.4 μm forward and reverse primers (IDT), 1.25 U Taq polymerase (Fermentas, USA) and milliQ water q.s. Final reaction volume of 25 μL was made with 5 μL of extracted fungal DNA (~0.5 ng). PCR assay was performed in a thermal cycler (Applied Bio system, USA) with the temperature profile: initial denaturation at 94°C for 5 minutes, followed by denaturation at 94°C for 30 seconds, primer annealing at 52°C for 30 seconds (ITS1 region) and 54°C (ITS2 region), strand elongation at 72°C for 30 seconds for 35 cycles, with the final elongation at 72°C for 10 minutes. Reaction mixture with 5 μL distilled water was used as a negative control and reaction mixture with DNA isolated from known isolates of Aspergillus flavus was used as a positive control in the PCR assay. Amplified PCR products were electrophoresed on 1.5% agarose gel and visualised under a gel documentation system (Syngene, USA).
Sequencing and sequence homology analysis: Amplified DNA bands for ITS1 and ITS2 regions were cut from the agarose gel and DNA was extracted using QIAquick Gel Extraction Kit (QIAGEN, USA) as per the manufacturer’s instructions. Nucleotide sequences of the purified DNA were determined commercially (Biolink, India) using primers ITS1 and ITS4 (sequences described earlier). Nucleotide sequences of ITS1 and ITS2 regions were searched for homology analysis with available sequences found in the Gene bank with NCBI BLAST computer program (NCBI, USA). Nucleotide sequences of both ITS1 and ITS2 regions from 31 culture isolates were submitted in the NCBI databank (NCBI, USA) (Nucleotide sequence accession numbers are given in results).
Results
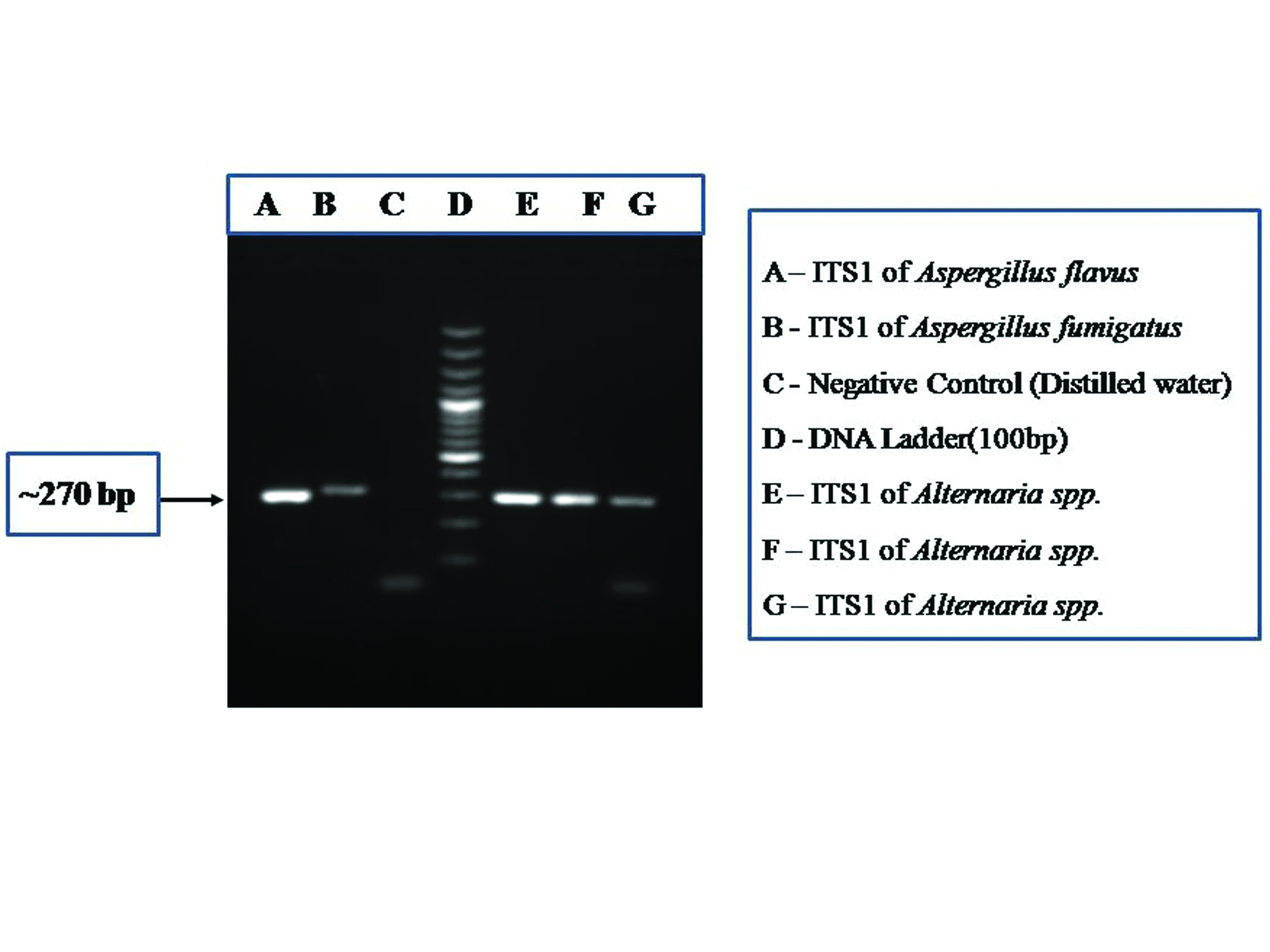
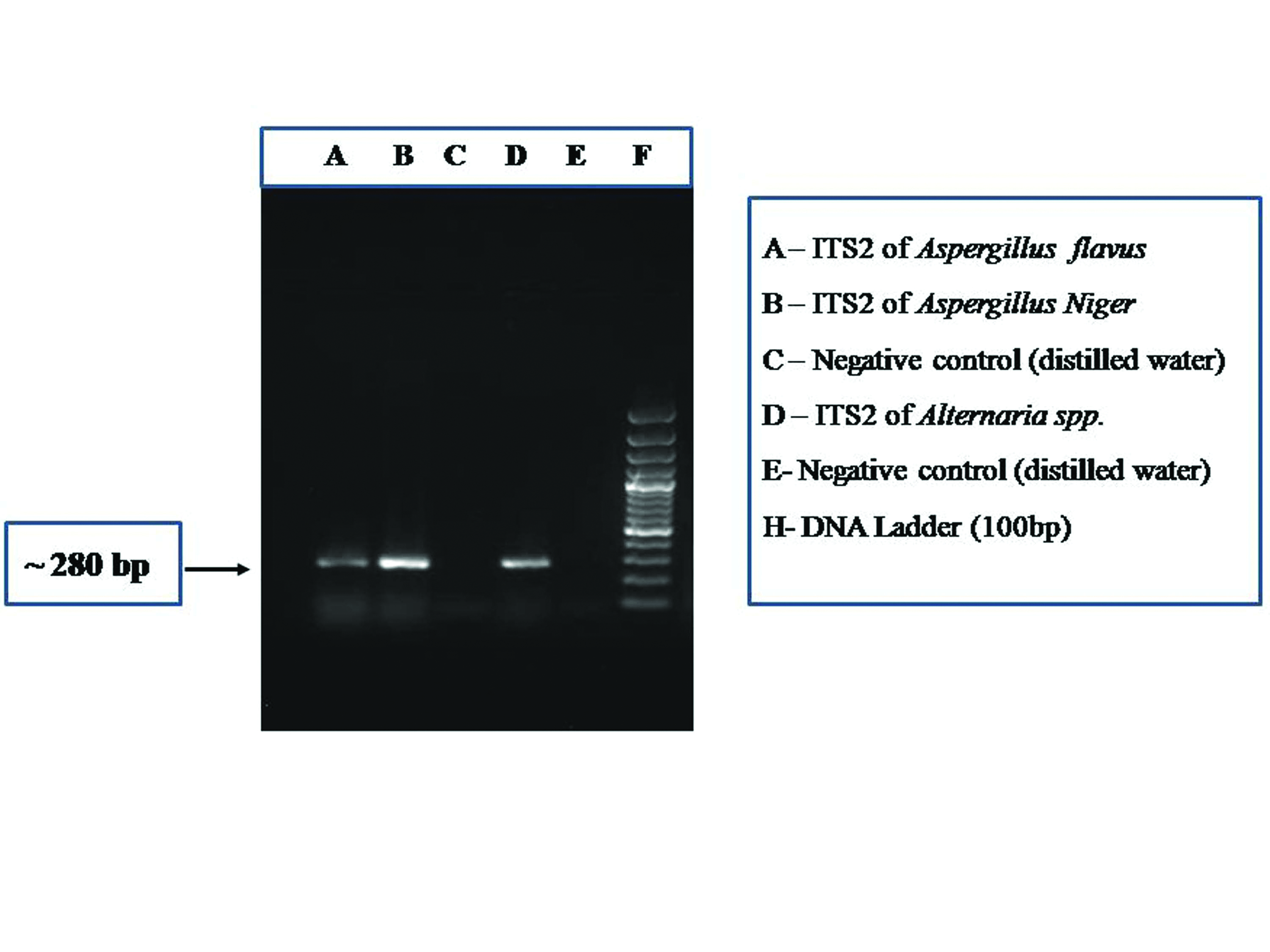
Of the 31 fungal culture isolates, visual and microscopic examinations could identify 22 isolates up to genus level and nine isolates up to species level [Table/Fig-1]. Amplified PCR products for the ITS1 and ITS2 regions when electrophoresed on 1.5% agarose gel shows bands of approximately 280 bp length when visualised with a gel documentation system [Table/Fig-2,3]. Nucleotide sequences submitted to the NCBI databank are now available with the accession numbers MF000891-MF000910 and MF033427-MF033435 for ITS1 region and MF000921-MF000940 and MF033438-MF033445) for ITS2 region [Table/Fig-1]. Of the 31 fungal culture isolates taken for sequencing to reach species level identification, eight were identified as Aspergillus flavus, one as Aspergillus fumigatus, four as Cladosporium cladosporioides, three as Simplicillium spp., three as Fusarium solani, one as Fusarium equiseti, three as Alternaria tenuissima, one as Alternaria alternata, two as Penicillium chrysogenum, one as Penicillium citrinum, one as Rhizopus microsporus, one as Rhizopus oryzae, one as Bipolaris sorokiniana and one as Chaetomium globosum. In five instances, fungal isolates identified by molecular methods were different from conventional methods [Table/Fig-1].
Details of 31 fungal isolates with accession numbers obtained from NCBI.
| S. No. of Patients | Nature of specimen | Fungus identified by conventional culture | Fungus identified by sequencing and NCBI-BLAST | Accession numbers obtained (NCBI) (ITS1 and ITS2) | Identity (%) | Accession numbers showing highest similarity |
|---|
| 1 | Corneal scraping | Acremonium spp. | Simplicillium spp. | MF000891 and MF000921 | 99% | KX020567, KX020563 |
| 2 | Corneal scraping | Aspergillus fumigatus | Aspergillus fumigatus | MF000892 and MF000922 | 99% | KU743889, JN226978 |
| 3 | Corneal scraping | Acremonium spp. | Simplicillium spp. | MF033427 and MF033438 | 99% | KT318874, KP184323 |
| 4 | Corneal scraping | Fusarium spp. | Fusarium solani | MF000893 and MF000923 | 99% | GQ451337, GQ121291 |
| 5 | Corneal scraping | Bipolaris spp. | Bipolaris sorokiniana | MF033428 (ITS1) | 98% | KU870641, KT310049 |
| 6 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF000894 and MF000924 | 99% | LN482517, LN482514 |
| 7 | Corneal scraping | Alternaria spp. | Chaetomium globosum | MF000895 and MF000925 | 99% | KU936228, KP281435 |
| 8 | Corneal scraping | Cladosporium spp. | Cladosporium cladosporioides | MF000896 and MF000926 | 96% | MF000907, MF000906 |
| 9 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF033439 (ITS2) | 99% | KR611594, JQ781721 |
| 10 | Corneal scraping | Fusarium spp. | Fusarium solani | MF033440 (ITS2) | 99% | KR527137, KY848498 |
| 11 | Invasive (Tissue) | Fusarium spp. | Fusarium equiseti | MF000897 and MF000927 | 100% | KR812230, KJ677237 |
| 12 | Corneal scraping | Penicillium spp. | Penicillium chrysogenum | MF000898 and MF000928 | 99% | MF000902, KX901289 |
| 13 | Corneal scraping | Alternaria spp. | Alternaria tenuissima | MF033429 (ITS1) | 100% | KR709011, KR912298 |
| 14 | Corneal scraping | Alternaria spp. | Alternaria tenuissima | MF000899 and MF000929 | 100% | HQ647307, KU508797 |
| 15 | Corneal scraping | Alternaria spp. | Alternaria alternata | MF033430 and MF033441 | 99% | MF040794, MF168401 |
| 16 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF000900 and MF000930 | 99% | KU561920, KU561919 |
| 17 | Corneal scraping | Acremonium spp. | Simplicillium spp. | MF000901 and MF000931 | 99% | KX020567, KX020563 |
| 18 | Invasive (Aspirate) | Penicillium spp. | Penicillium chrysogenum | MF000902 and MF000932 | 99% | KU982597, JF807949 |
| 19 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF000903 and MF000933 | 99% | KP296143, KX345284 |
| 20 | Corneal scraping | Alternaria spp. | Alternaria tenuissima | MF000904 and MF000934 | 100% | LT799975, KX064997 |
| 21 | Corneal scraping | Fusarium spp. | Fusarium solani | MF000905 and MF000935 | 99% | GQ451337, GQ121291 |
| 22 | Corneal scraping | Cladosporium spp. | Cladosporium cladosporioides | MF000906 and MF000936 | 99% | JN227029, KX960912 |
| 23 | Corneal scraping | Cladosporium spp. | Cladosporium cladosporioides | MF000907 and MF000937 | 99% | KP689250, KF876823 |
| 24 | Invasive (Tissue) | Aspergillus flavus | Aspergillus flavus | MF033431 and MF033442 | 99% | MF163443, MF120213 |
| 25 | Invasive (Tissue) | Rhizopus spp. | Rhizopus microsporus | MF033432 (ITS1) | 99% | KJ408570, KM527225 |
| 26 | Invasive (Tissue) | Aspergillus flavus | Aspergillus flavus | MF000908 and MF000938 | 99% | HM560052, HM560051 |
| 27 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF033433 and MF033443 | 100% | MF033444, MF033443 |
| 28 | Invasive (BAL) | Penicillium spp. | Penicillium citrinum | MF000909 and MF000939 | 99% | KY754577, KX363454 |
| 29 | Invasive (Tissue) | Mucor spp. | Rhizopus oryzae | MF000910 and MF000940 | 99% | KX685359, FJ433877 |
| 30 | Corneal scraping | Aspergillus flavus | Aspergillus flavus | MF033434 and MF033444 | 100% | MF033444, MF033443 |
| 31 | Corneal scraping | Cladosporium spp. | Cladosporium cladosporioides | MF033435 and MF033445 | 100% | KJ728689, KJ410037 |
PCR assay showing the amplification of ITS1 region (~270 bp) of fungus.
PCR assay showing the amplification of ITS2 region (~280 bp) of fungus.
Nucleotide sequences of Aspergillus flavus (n=8) from conventional culture share 100%/99% identity with Aspergillus flavus by sequencing and NCBI-BLAST (accession numbers: LN482517, LN482514, LN482513, KR611594, JQ781721, FJ011545). Similarly, Aspergillus fumigatus (n=1) from conventional culture shares 99% identity with Aspergillus fumigatus (accession numbers: KU743889, JN226978, JX469423). All three Cladosporium spp. share 100%/99% identity with Cladosporium cladosporioides (KJ728689, KP689250, KX610162). All the three Acremonium spp. share 99% identity with Simplicillium spp. (KX020567, KX020563, KP184324). Of the four Fusarium spp., three share 99% identity with Fusarium solani (GQ451337, GQ121291, KX381129) and one shares 100% identity with Fusarium equiseti (KR812230, KJ677237, KJ677236). Of the five Alternaria spp., three share 100% identity with Alternaria tenuissima (HQ647307, KU508797, KX064997), one shares 99% identity with Alternaria alternata (MF040794, MF168401, MF167641) and one shares 99% identity with Chaetomium globosum (KU936228, KP281435, JX406510). Of the three Penicillium spp., two share 99% identity with Penicillium chrysogenum (MF000902, KX901289, KU743900) and one shares 99% identity with Penicillium citrinum (KY754577, KX363454, KX363446). Rhizopus spp. (n=1) shares 99% identity with Rhizopus microsporus (KJ408570, KM527225, KM527224). Mucor spp. (n=1) shares 99% identity with Rhizopus oryzae (KX685359, FJ433877, KY244030) and Bipolaris spp. (n=1) shares 98% identity with Bipolaris sorokiniana (KU870641, KT310049, KF922886).
Discussion
Keratomycosis is one of the most frequently encountered ophthalmic infections in tropical and sub tropical countries like India, caused by a variety of fungal species including opportunistic fungal pathogens and in some instances by these, previously classified as contaminant fungi [28]. IFIs are the leading causes of morbidity and mortality in immunocompromised patients [9,10]. Accurate identification of fungi to the species level holds an importance in developing countries like India, where fungal infections are more common and correct choice of antimicrobial therapy requires fungal infections to be distinguished from other microbial aetiology. As identification of the causative fungi with conventional culture needs more than a week, rapid and high throughput molecular diagnostic tools like PCR assay and sequencing are very helpful for species level identification either from culture isolates or directly from clinical specimens for correct treatment [26]. In this pilot study, we have identified the pathogenic fungi to species level from culture positive clinical isolates to guide better therapy for the management of infections. Some fungal isolates could be fully identified by conventional methods such as visual and microscopic morphological examinations. Correct identification is important because treatment varies among different genus of fungi and in some cases from species to species, as relatively high Minimum Inhibitory Concentrations (MICs) of amphotericin B and itraconazole are needed for F. solani compared to F. oxysporum [29].
Fungal species identification is based on the variation in nucleotide sequences of the variable ITS1 and ITS2 regions of ribosomal DNA (rDNA). As 28S, 18S and 5.8S rRNA are the conserved regions of fungal ribosomal DNA, these are often used for designing forward and reverse primers to amplify the inner variable regions in PCR assay. ITS1 and ITS2 regions can be amplified jointly with forward primer ITS1 and reverse primer ITS4, which is about 560 bp in length, but it has some limitations due to “chimera formation”. To avoid such type of limitations, these targets (ITS1 and ITS2) can be amplified separately for species level identification [26,30,31].
Among Aspergillus species, A. flavus is the predominant fungi causing mycotic keratitis and second only to A. fumigatus as a cause of human invasive infections [32,33]. In this pilot study, it was also reported that, of the total seven Aspergillus spp. isolates from mycotic keratitis patients, six were A. flavus and only one was A. fumigatus which corroborates with the previous studies.
The species of Simplicillium is considered as coincidental opportunistic pathogen to humans and animals as it occurs in a wide range of ecological niches, such as soil, mushroom, diseased plant tissue, rust, nematode, dog tissue and human nails [34]. Acremonium spp. is morphologically similar to Simplicillium spp. where the isolates do not produce their distinctive macroconidia. Some of the species of Simplicillium such as Simplicillium obclavatum, originally described as Acremonium obclavatum, which makes it difficult to differentiate between them by conventional methods [35,36]. Therefore, genus level identification of both the fungus is difficult exclusively on morphological characteristics and there is always a possibility of misidentification. Hence, molecular identification may be performed as a complementary test to avoid such type of misdiagnosis. In this study, phenotypically identified all three isolates of Acremonium spp. was identified as Simplicillium spp. by molecular methods.
Cladosporium spp. was considered as a coincidental opportunistic pathogen, causing mycotic keratitis and some cutaneous and subcutaneous infections [37]. Cladosporium cladosporioides is a very rare pigmented fungi with few reported cases of keratomycosis [2]. In some case reports from Asian and African countries, corneal keratomycosis resulting from Cladosporium cladosporioides had been reported [38-40]. In the present study, we found four isolates of Cladosporium cladosporioides with PCR assay and sequencing from mycotic keratitis patients which were reported as Cladosporium spp. with traditional mycological diagnostic method.
A study from India reported the incidence of Alternaria spp. in mycotic keratitis patients as 3.3-10.4% [41]. Among all Alternaria spp., A. alternata and A. tenuissima are more common in ocular infections [41]. In the present study, we found one isolate of A. alternata and three isolates of A. tenuissima, which corroborates with the previous studies. Alternaria spp. and Chaetomium spp. both belong to dematiaceous moulds, so they can be misidentified by conventional methods. In the present study, one isolate which was identified as Alternaria spp. by conventional methods, later characterised as Chaetomium globosum by molecular methods. Association of Chaetomium globosum was already reported with keratomycosis [42].
Species level identification of Zygomycetes by standard mycological methods always remains a difficult and time-consuming task, which requires an expertise that is restricted to few reference laboratories. DNA typing to differentiate between the species among Zygomycetes is particularly important in those cases where these fungi differ in their susceptibility to antifungal drugs. The two most common pathogenic species of Rhizopus i.e., R. oryzae and R. microsporus, which are difficult to differentiate phenotypically, could be clearly differentiated from each other genotypically, as their sequences showed only 70% similarity [43]. In the present study, we found one Rhizopus microsporus from tissue specimen, which was earlier reported as Rhizopus spp. Another invasive fungal isolate which was misidentified as Mucor spp. by conventional methods was identified as Rhizopus oryzae with molecular diagnosis. These molecular typing results hold importance in differentiating between different species of Zygomycetes to start an appropriate antifungal therapy. Hence, molecular diagnosis is useful for species identification within Zygomycetes from culture isolates.
Studies from South India and central China reported Fusarium spp. as the most common cause of mycotic keratitis; among which F. solani is the most predominant one [28,44]. Few cases of F. equiseti infections were also reported in some parts of the world [45,46]. In the present study, we had reported three F. solani and one F. equiseti isolates from fungal infections by nucleotide sequencing, which suggests that, F. solani is most prevalent among all Fusarium spp. causing eye infections.
This study presented a standardised rapid and high throughput technique (PCR assay) for the detection and identification of fungi in ocular and invasive samples. It consists of a sensitive, precise, rapid but relatively expensive method of identification of fungi based on the amplification of ITS1 and ITS2 regions and sequencing. There are several species within a genus that differ in their susceptibility to antifungal drugs, in those instances molecular typing by PCR assay can give guidance for better therapy to manage such infections. Molecular assays are neither been widely used nor widely available. There is a need for widespread use of molecular assays for correct identification, so that some fungal isolates will not be discarded as contaminants. Misidentification of rare isolates can be avoided by using molecular techniques.
Limitation
The present study included culture isolates of filamentous fungi only. IFIs due to Candida spp. could also have been included.
Conclusion
Molecular diagnosis with PCR assay can be an important complement to conventional culture methods (gold standard) and may constitute a rapid and high throughput means of fungal identification, which is becoming very important in clinical mycology. As the cost of PCR assay to diagnose infections generally exceeds that of conventional culture, which limits its widespread use, especially in low income countries like India, it should be implemented at least in reference centers for a definitive diagnosis and treatment.