Introduction
Multiple Myeloma (MM) is a cytogenetically heterogeneous haematologic malignancy characterised by uncontrolled proliferation of clonal plasma cells within the bone marrow. TP53 gene inactivation is considered as an independent prognostic marker and patients harbouring these mutations are usually resistant to standard therapy.
Aim
To determine the frequency of TP53 gene mutations in exons 4 to 9 and the distribution of Arg72Pro polymorphism in exon 4 in newly diagnosed multiple myeloma patients.
Materials and Methods
Mutation analysis of genomic DNA from unsorted bone marrow aspirates of 30 patients (10 showed TP53 deletion by interphase FISH) and from peripheral blood lymphocytes of 30 healthy control individuals was performed by direct sequencing of amplified products using self-designed primers. The codon 72 polymorphism was studied using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) technique in an additional 108 MM patients and 70 healthy controls.
Results
TP53 gene alterations were recorded in six patients (20%) and three of them showed two or more changes. No alterations were observed in exons 5, 7 and 9 in myeloma patients. Four mutations in codons c.284C>T (exon 4), c.641A>G (exon 6), c.787A>G and c.808T>G (exon 8) and three intronic variants c.672+48G>A (intron 6), c.782+72C>T and c.782+92T>G (intron 7) were seen only in the patient group. The variants c.108G>A (exon 4), c.672+62A>G (intron 6) and c.993+12T>C (intron 9) were observed in both groups. Three patients died within six months of diagnosis. The genotype and allele frequencies for Arg72Pro polymorphism were similar in the patient and in the control groups.
Conclusion
The presence of TP53 mutations denoted a poor prognosis while the TP53 Pro72Arg polymorphism is not associated with increased risk for MM.
Introduction
Multiple Myeloma (or plasma cell myeloma) belongs to a group of paraproteinaemias that progresses through clinically distinct Monoclonal Gammopathy of Unknown Significance (MGUS), asymptomatic or Smouldering Multiple Myeloma (SMM), (symptomatic) MM and Plasma Cell Leukaemia (PCL). MM is characterised by translocations involving the Immunoglobulin Heavy Chain (IGH) gene leading to oncogenic activation, hyperdiploidy through gain of one or more odd-numbered chromosomes 3, 5, 7, 9, 11, 15, 19 and 21, copy number anomalies, gene mutations, epigenetic changes and miRNA abnormalities. Further, complex cytogenetic abnormalities (≥3 abnormalities), monosomy 13/del (13q) or monosomy 17/del(17p) are usually considered as markers of adverse prognosis [1-4]. Specific genetic abnormalities may probably be ‘driver’ events determining clonal selection and disease evolution. The heterogeneity from acquisition of additional genetic “hits” over time then confers a survival advantage as determined by drug resistance and tumour progression [5].
The TP53 tumour suppressor gene located at 17p13 encodes a nuclear phosphoprotein which is implicated in the regulation of progression through cell cycle and control of apoptosis. The gene acts in response to stress by inducing target genes that regulate cell-cycle arrest by blocking G0/G1-S transition, DNA repair, apoptosis or changes in metabolism [6-8]. TP53 mutations and polymorphisms have been reported in about one-half of all human tumours and involves all the coding exons, more frequently the exons 4 to 9 which encode the DNA-binding domain of the protein [9,10]. The frequency of TP53 mutations increased with disease stage being about 13% at diagnosis of MM (7/52 unselected) and 43% in advanced MM or PCL (7/16 cases). No mutations were detected in patients with MGUS or SMM [11].
The codon 72 polymorphism rs1042522 (Arg72Pro) in TP53 gene is the most widely studied polymorphism in cancers and conflicting results have been reported on its association with susceptibility to haematological malignancies [12]. The Arg variant has been demonstrated to be more potent in initiating apoptosis while Pro variant is associated with arrest of cell cycle and DNA repair [13-17]. There was no significant difference in the frequency of the polymorphism between Japanese MM patients and healthy individuals in a study carried out in Japan [18]. Due to inadequate information on TP53 alterations in MM, this study was aimed to determine the frequency of TP53 mutations in exons 4 to 9 and the distribution of codon 72 polymorphism in newly diagnosed MM patients.
Materials and Methods
A total of 215 MM patients were registered at the Department of Centralised Molecular Diagnostics, Apollo Hospitals, Chennai, India during the period of July 2012 to December 2015. Prior approval was taken for the present study from the Ethics Committee of Apollo Hospitals, Chennai. Heparinised bone marrow aspirates collected after having obtained written informed consent from these patients were processed for interphase fluorescence in situ hybridisation (iFISH) using 7-probe panel for MM. DNA was isolated from EDTA-coated bone marrow aspirates of 15 patients who exhibited del(17p) and 20 cases which were negative for the deletion by iFISH using a TP53-specific probe. Genomic DNA was also isolated from peripheral blood lymphocytes of 30 healthy individuals using the same phenol-chloroform isoamyl alcohol method [19]. The DNA quality was found to be bad in five patients who had del(17p) upon qualitative analysis using Nanodrop Spectrophotometer (Thermo Scientific, Germany) and by running on 0.8% agarose gel [19].
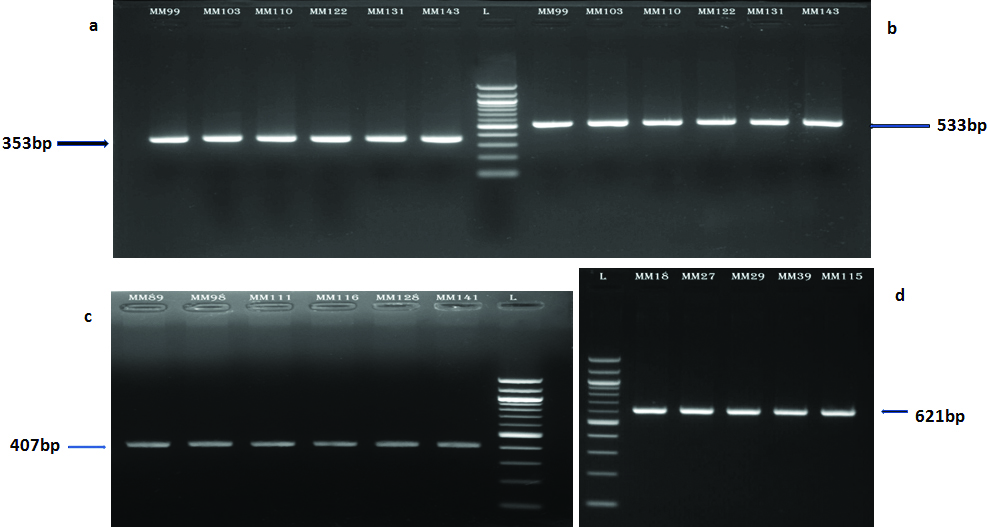
The DNA samples were amplified by Polymerase Chain Reaction (PCR) and analysed for the presence of mutations in exons 4 to 9 of the TP53 gene by direct sequencing. The PCR products for exon 4 (353bp), exons 5-6 (533bp), exon 7 (407bp) and exons 8-9 (621bp) [Table/Fig-1] were obtained using self-designed primers and were purified employing Qiagen purification kit. The sequences for the primers were as follows:
Agarose gel electrophoresis of PCR products of: a) Exon 4;
b) Exons 5-6; c) Exon 7; and d) Exons 8-9 of TP53 gene.
PCR-Polymerase Chain Reaction
1) Exon 4 - F-5’TCTGACTCTTTTCACCC3’, R-5’CCAGGCATTG AAGTCATGGAAG3’;
2) Exons 5-6 - F-5’CTTTCAACTCTGTCTCCTTCC3’, R-5’TCATGGGGTTATAGGGAGGTC3’;
3) Exon 7 - F - 5’CGACAGAGCGAGATTCCATC3’, R-5’GTTAAGAGGTCCCAAAGCCAGAG3’; and
4) Exons 8 - 9 - F - 5’GCTTTGGGACCTCTTAACCT3’, R-5’GCTACAACCAGGAGCCATTG3’.
PCR conditions comprised of initial denaturation for five minutes at 95°C followed by 35 cycles of denaturation at 94°C for 30 seconds and extension at 72°C for 30 seconds. Annealing was carried out for 30 seconds at 61°C for exons 4 and 7, 62°C for exons 5-6 and 60°C for exons 8-9. A final extension of seven minutes at 72°C was given. The PCR products were visualised (before and after purification) on 1.5% agarose gel and recorded using gel documentation system. The purified products were commercially sequenced at Madras Diabetic Research Foundation, Siruseri, Chennai, India. The electropherograms were checked using Finch TV and the sequences obtained for the selected exons in 30 cases each of MM and controls were compared employing Bioedit software through multiple sequencing alignment with the reference sequence available in the IARC TP53 database [20].
The Arg72Pro (rs1042522) polymorphism in TP53 gene was studied using PCR-RFLP technique in 108 patients with MM (who did not exhibit del (17p) using iFISH and in 70 heathy controls besides those individuals examined by direct sequencing). A small quantity (5 μL) of the PCR products were visualised on 1.5% agarose gel and then the remaining 20 μL were digested with the restriction enzyme BstUI (New England Biolabs) at 60°C for five hours following the manufacturer’s instructions. The digested products were resolved on 2% agarose gel. The sizes of the digested amplicons were determined using 100 bp ladder. The fragments were visualised under UV light and documented (Syngene Bioimaging Pvt. Ltd., Haryana, India).
Statistical Analysis
Gene frequencies were calculated by gene counting method. Deviation from Hardy-Weinberg equilibrium was calculated by Fisher’s exact two-sided test. Odds Ratio (OR) for the allele and genotype frequencies for the Arg72Pro polymorphism in exon 4 of TP53 gene by direct sequencing or RFLP were computed using 2x2 contingency tables. The genotypic test for association between each Single Nucleotide Polymorphism (SNP) and MM was done based on three models (Codominant, dominant and recessive). A p-value of <0.05 was considered to be significant.
Results
Screening for TP53 mutations by direct sequencing in 30 MM patients (10 who were positive and 20 were negative for TP53 deletion by iFISH) and 30 healthy controls revealed no changes in exons 5, 7 and 9. Six patients (20%) showed gene alterations, mostly represented by a heterozygous single nucleotide change, and three of them showed two or more changes. Four alterations within the codons c.284C>T (exon 4), c.641A>G (exon 6), c.787A>G and c.808T>G (exon 8) and three intronic variants c.672+48G>A (intron 6), c.782+72C>T and c.782+92T>G (intron 7) were seen only among MM patients. The variants c.108G>A (exon 4), c.672+62A>G (intron 6) and c.993+12T>C (intron 9) were observed in both the cohorts [Table/Fig-2]. All these six patients presented with typical clinical complications characteristic of MM with bone marrow infiltration (>60% plasma cells). Four of them (MM 110, MM 131, MM 138, MM 151) showed hemizygous TP53 deletion by iFISH. The survival duration was less than six months from the time of diagnosis in three patients harbouring TP53 mutations (MM 98, MM 103, MM 151) while the remaining three were lost to follow up. This study also revealed the substitution c.903A>G resulting in the synonymous alteration p.P301P in the control MC26 individual.
Spectrum of mutations in exons 4 to 9 of TP53 gene detected by direct sequencing in this study.
| Case ID/ Age/Sex | Plasma cells (%) | Exon/Intron | g_Description | c_Description | Amino acid change | Status |
|---|
| MM 98/45/F | 60 | Exon 4 | g.11515C>T | c.284C>T | p.S95F | Died (three months) |
| Exon 8 | g.13767A>G | c.787A>G | p.N263S |
| MM 103/51/M | 67 | Intron 7 | g.13491C>T | c.782+72C>T | - | Died (two months) |
| Intron 7 | g.13511T>G | c.782+92T>G | - |
| MM 110*/67/M | 64 | Exon 4 | g.11339G>A† | c.108G>A† | p.P36P | Lost to follow up |
| Intron 6 | g.12789G>A | c.672+48G>A | - |
| Intron 9 | g.14077T>C† | c.993+12T>C† | - |
| MM 131*/49/F | 86 | Exon 6 | g.12710A>G | c.641A>G | p.H214R | Lost to follow up |
| MM138*/77/M | 70 | Exon 8 | g.13790T>G | c.808T>G | p.F270L | Lost to follow up |
| MM151*/51/M | 83 | Intron 6 | g.12803A>G† | c.672+62A>G† | - | Died (five months) |
*positive for TP53 deletion by interphase FISH;
†changes seen in both patients and controls
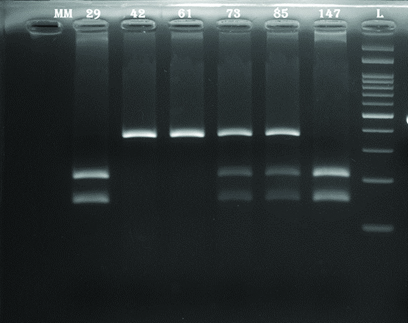
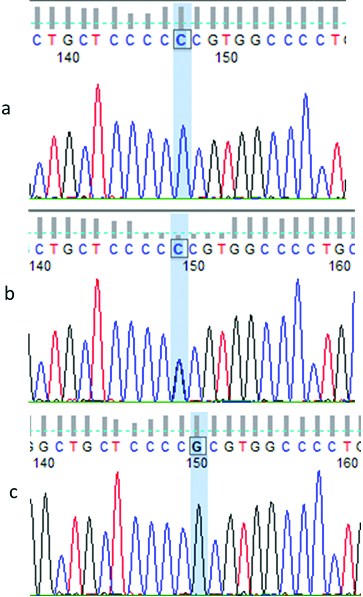
The c.215 G>C (Arg72Pro) (rs1042522) polymorphism in exon 4 of TP53 gene was studied using direct sequencing (in 30 each of MM patients and controls) and by PCR-RFLP (in 108 MM patients and 70 control individuals) methods. This study included 87 male patients with a mean age of 57.9±10.5 (range: 33-78 years) and 51 females having a mean age of 57.7±10.4 (range: 31-81 years). The ‘C’ allele eliminates a site for the enzyme to produce a single fragment of 353 bp (as the undigested product) in the Pro/Pro homozygous individual while two fragments of sizes 206 bp and 147 bp in the Arg/Arg homozygous individual and three fragments 353 bp, 206 bp and 147 bp in the Arg/Pro heterozygote are observed [Table/Fig-3]. The partial electropherograms showing the three genotypes CC, GC and GG are shown in [Table/Fig-4]. A total of 104 MM patients (75.36%) harboured the p.R72P polymorphism in comparison with 81% of the non-cancer controls. The distribution of the genotype frequencies was consistent with the Hardy-Weinberg equilibrium in both groups (p-value=0.74 and 0.15 respectively). The frequencies of both the alleles G and C were similar in the patient (0.49 and 0.51) and in the control (0.475 and 0.525) groups. The Pro/Pro genotype was observed in 34 patients (24.6%), Pro/Arg in 73 (52.9%) and Arg/Arg in 31 (22.5%) [Table/Fig-5]. Statistical analysis using dominant, codominant and recessive genetic models revealed that the TP53 Arg72Pro polymorphism was not associated with increased risk for MM.
2% Agarose gel electrophoresis of Pro72Arg (rs1042522) polymorphism in exon 4 of TP53 gene showing RFLP patterns up on digestion using BstUI -Lanes 1 and 6-Arg/Arg genotype (206 bp, 147 bp) Lanes 2 and 3-Pro/Pro genotype (353 bp); Lanes 4 and 5–Pro/Arg genotype (353 bp, 206 bp, 147 bp); Lane 7-100 bp ladder.
RFLP-Restriction Fragment Length Polymorphism
Partial electropherograms showing TP53 codon 72 polymorphism: a) CC homozygous genotype (Pro/Pro); b) CG heterozygote (Pro/Arg); c) GG homozygous genotype (Arg/Arg).
Genotypic and allelic distribution of Pro72Arg polymorphism in TP53 gene in multiple myeloma patients and control individuals.
| Models | Cases (n=138) (%) | Controls (n=100) | OR (95% CI) | p-value |
|---|
| Codominant | | |
| GG | 34 (24.64) | 19 | 1.00 | |
| GC | 67 (48.55) | 57 | 1.52 (0.78-2.99) | 0.22 |
| CC | 37 (26.81) | 24 | 1.16 (0.54-2.50) | 0.71 |
| Dominant | | |
| GG | 34 (24.64) | 19 | 1.00 | |
| GC+CC | 104 (75.36) | 81 | 1.39 (0.74-2.66) | 0.31 |
| Recessive | | |
| GG + GC | 101 (73.19) | 76 | 1.00 | |
| CC | 37 (26.81) | 24 | 0.86 (0.47-1.56) | 0.63 |
| Allele contrast | | |
| G | 135 | 95 | 1.00 | |
| C | 141 | 105 | 1.06 (0.73-1.53) | 0.76 |
| HWE χ2 | 0.11 | 2.04 | | |
| HWE p-value | 0.74 | 0.15 | | |
OR-Odds Ratio, CI-Confidence Interval
Discussion
Deletion of the short arm of chromosome 17, more specifically 17p13 which is the site of the tumour suppressor gene TP53 commonly occurs as a secondary cytogenetic abnormality and is associated with aggressive disease, the presence of extramedullary disease and shorter survival [3,4]. The TP53 gene encodes the p53 phosphoprotein, and is referred to as ‘the guardian of the genome’ due to its key role in cell cycle regulation, DNA repair and apoptosis. This gene is found to be mutated in more than 50% of all cancers [21]. TP53 inactivation by either deletion or mutation is a rare event in MM being reported only in the late stages of disease progression. The prevalence of mutations is about 5% at diagnosis, 20–40% in advanced MM or PCL and in >60% of human myeloma cell lines [22]. Chng WJ et al., detected TP53 mutations in only 3% of 268 newly diagnosed MM patients [23]. Further, a greater proportion of patients with del(17p) exhibited mutations in comparison with those lacking it [23,24]. Four of the six patients (MM 110, 131, 138, 151) with TP53 mutations in the present study showed del(17p) by iFISH. A total of five mutations in the codons (one polymorphic) and five in the intronic regions (two polymorphic) were observed upon screening of 30 patients with MM. All these changes have not been reported in MM earlier to our knowledge. Mutations were also not found within the six ‘hotspot’ codons 175, 245, 248, 249, 273 and 282 that account for 30% of all TP53 mutations in cancers [10]. Further, the presence of TP53 mutations had an extremely negative prognostic significance being associated with a poor survival of less than six months. This is in accordance with existing reports on MM [23] and in other haematological cancers such as chronic lymphocytic leukemia [25] and acute myeloid leukemia [26,27]. The authors had even suggested that patients with TP53 mutations should be considered for alternative and novel therapies due to their poor response to standard therapeutic approaches. The selection of somatic TP53 mutations during tumourigenesis results from both dominant negative and gain-of-function mechanisms [10].
The transition A to G at codon 214 in exon 6 resulting in the substitution p.H214R was observed in the patient MM 131 who had exhibited TP53 deletion using iFISH. The p.H214R mutation has been reported earlier in two cases of prostate cancer and in a case of primary colon cancer [7,28]. Yang Y et al., observed the A→G transition at codon 263 in exon 8, c.787A>G, to be a frequent hotspot for p53 mutations in their evaluation of 34 patients who underwent radical resection for their primary colorectal cancers and subsequently partial hepatectomy for hepatic metastases [29]. This transition was expressed as a heterozygous change in the patient MM 98 who did not respond and died within three months. The same individual also expressed another missense mutation c.284C>T which has been described in skin tumours [20]. The patient MM 110 who carried the del(17p) by iFISH also had the codon 36 transition c.108G>A causing a synonymous p.P36P change. This polymorphism was found in a healthy individual in this study. In support of our finding, Takahashi Y et al., had described this previously known transition G→A in codon 36 in both the primary tumour and its adjacent normal mucosa [28]. A missense mutation (T–>G) in exon 8, which resulted in the conversion of phenylalanine to cysteine at codon 270 was seen in another patient (MM 138). This transversion c.808T>G was reported earlier in a 37-year-old male with Astrocytoma Grade II. Further, a correlation of the occurrence of p53 mutations in gliomas with age was observed, being rare among paediatric age groups, high in young adults and low among the older age group [30]. The substitution c.903A>G leading to the silent p.P301P change and seen in a control sample had been described earlier in oesophageal tumours [20].
The common p53 polymorphisms include codon 72 (c.215C>G; p.R72P), deletion of 16bp in intron 3 (c.96+41_96+56del16) and c.672+62A>G polymorphisms [12]. While the two SNPs were only detected the deletion was not registered in this study. All three genotypes for the c.672+62A>G polymorphism in intron 6 were present in both cohorts. However, there seemed to be no association with increased risk for MM (OR=2.05; p=0.62). The patient MM 110 was heterozygous for this SNP and another polymorphism located 14 bp upstream, c.672+48G>A. Berggren P et al., observed the polymorphism c.782+72C>T within intron 7 of the TP53 gene in 15% of their 159 breast cancer patients and 171 non-cancer individuals [31]. It was also seen that all these individuals exhibited another polymorphism c.782+92T>G situated 20 bp downstream. Prosser J and Condie A had earlier described the same two linked polymorphisms in 10% of their breast cancer patients and in 13% of the controls [32]. It was of interest to find both these SNPs in the patient negative for del(17p) by iFISH (MM 103) in this study. The patient died two months after diagnosis. Palmero EI et al., assigned a possibly pathogenic significance to the c.782+92T>G polymorphism in their patients fulfilling Li-Fraumeni-like syndrome criteria [33]. The other common polymorphisms reported were c.993+12T>C and c.108G>A. The nucleotide replacement c.993+12T>C in intron 9 was noted as polymorphism occurring in a single individual each from the case (MM 110) and control groups. This change was however, reported in a single patient out of 32 patients with early-onset colorectal cancer screened for TP53 mutations [34]. Subsequently, Zhunussova G et al., found three cases of advanced Colorectal Cancer (CRC) carrying this mutation [35]. However, nothing was stated about the pathological significance of this intronic mutation as the percentage of TP53 mutations among advanced CRC patients from Kazakhstan was very low (3.4%). None of the intronic mutations detected in the present study occurred at a splice site or created a new splice site. The pathological significance of these intronic polymorphisms needs further investigation.
Studies on Arg72Pro polymorphism (rs1042522) at codon 72 in exon 4 of the TP53 gene have reported an association of the Arg allele with increased risks for colorectal, breast and lung cancers and the Pro allele with lung, thyroid, breast and cervical cancers [18]. A meta-analysis showed a significantly increased risk for non-Hodgkin lymphoma in all subjects with p53 Arg72Pro polymorphism under the dominant model. However, no significant association was found between this polymorphism and leukaemia risk [12]. A recent study from India on advanced cervical cancer patients did not reveal any significant difference in the Pro/Pro (23.3%), Arg/Pro (49.5%) and Arg/Arg (27.2%) genotype frequencies with respect to patients overall survival and recurrence-free survival [36]. In contrast, Rao AKDM et al., demonstrated the Pro/Pro genotype in association with tobacco chewing habit to confer an increased risk to oral cancer in patients of South Indian origin [37]. The genotypic analysis of this c.215G>C polymorphism did not present a significantly different allele distribution in the patient compared to the control samples suggesting that it is not associated with an increased risk for MM and this was in accordance with earlier reports [18,38]. However, a strong association of Pro allele with earlier relapse and shorter overall survival was suggested to be due to the resistance of these cells to induction of apoptosis by thalidomide therapy [18]. This is the first report on TP53 gene alterations in patients with MM from India, to the best of our knowledge.
Limitation
Extended studies on larger number of MM patients in different disease stages and subsequently, on patients with other haematological cancers are essential to determine the entire spectrum of variants in both coding and intronic regions of the TP53 gene as well as to understand the biological effect of these changes in tumour progression.
Conclusion
The present study confirms the association of TP53 mutations with a poor prognostic outcome in patients with MM and stresses the need for inclusion of TP53 mutation testing in the initial work-up. The TP53 Pro72Arg polymorphism however, did not correlate with an increased risk for MM.
*positive for TP53 deletion by interphase FISH;
†changes seen in both patients and controls
OR-Odds Ratio, CI-Confidence Interval
[1]. Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma developmentCancer Cell 2010 18(4):367-81.10.1016/j.ccr.2010.09.00520951946 [Google Scholar] [CrossRef] [PubMed]
[2]. Morgan GJ, Walker BA, Davies FE, The genetic architecture of multiple myelomaNat Rev Cancer 2012 12(5):335-48.10.1038/nrc325722495321 [Google Scholar] [CrossRef] [PubMed]
[3]. Rajan AM, Rajkumar SV, Interpretation of cytogenetic results in multiple myeloma for clinical practiceBlood Cancer J 2015 5(10):e36510.1038/bcj.2015.9226517360 [Google Scholar] [CrossRef] [PubMed]
[4]. Talley PJ, Chantry AD, Buckle CH, Genetics in myeloma: genetic technologies and their application to screening approaches in myelomaBr Med Bull 2015 113:15-30.10.1093/bmb/ldu04125662536 [Google Scholar] [CrossRef] [PubMed]
[5]. Fairfield H, Falank C, Avery L, Reagan MR, Multiple myeloma in the marrow: pathogenesis and treatmentsAnn N Y Acad Sci 2016 1364(1):32-51.10.1111/nyas.1303827002787 [Google Scholar] [CrossRef] [PubMed]
[6]. Vogelstein B, Lane D, Levine AJ, Surfing the p53 networkNature 2000 408:307-10.10.1038/3504267511099028 [Google Scholar] [CrossRef] [PubMed]
[7]. Agell L, Hernandez S, de Muga S, Lorente JA, Juanpere N, Esgueva R, KLF6 and TP53 mutations are a rare event in prostate cancer: distinguishing between Taq polymerase artifacts and true mutationsModern Pathology 2008 21:1470-78.10.1038/modpathol.2008.14519020536 [Google Scholar] [CrossRef] [PubMed]
[8]. Reisman D, Takahashi P, Polson A, Boggs K, Transcriptional regulation of the p53 tumour suppressor gene in S-Phase of the cell-cycle and the cellular response to DNA damageBiochemistry Research International 2012 :80893410.1155/2012/80893422830025 [Google Scholar] [CrossRef] [PubMed]
[9]. Soussi T, Wiman TG, Shaping genetic alterations in human cancer: the p53 mutation paradigmCancer Cell 2007 12:303-12.10.1016/j.ccr.2007.10.00117936556 [Google Scholar] [CrossRef] [PubMed]
[10]. Rivlin N, Brosh R, Oren M, Rotter V, Mutations in the p53 tumour suppressor gene: important milestones at the various steps of tumourigenesisGenes and Cancer 2011 2(4):466-74.10.1177/194760191140888921779514 [Google Scholar] [CrossRef] [PubMed]
[11]. Neri A, Baldini L, Trecca D, Cro L, Polli E, Maiolo AT, p53 Gene mutations in multiple myeloma are associated with advanced forms of malignancyBlood 1993 81(1):128-35. [Google Scholar]
[12]. Weng Y, Lu L, Yuan G, Guo J, Zhang Z, Xie X, p53 codon 72 polymorphism and haematological cancer risk: an update meta-analysisPLoS ONE 2012 7(9):e4582010.1371/journal.pone.004582023029260 [Google Scholar] [CrossRef] [PubMed]
[13]. Dumont P, Leu JI, Della Pietra III AC, George DL, Murphy M, The codon 72 polymorphic variants of p53 have markedly different apoptotic potentialNat Genet 2003 33:357-65.10.1038/ng109312567188 [Google Scholar] [CrossRef] [PubMed]
[14]. Pim D, Banks L, p53 polymorphic variants at codon 72 exert different effects on cell cycle progressionIntl J Cancer 2004 108(2):196-99.10.1002/ijc.1154814639602 [Google Scholar] [CrossRef] [PubMed]
[15]. Siddique M, Sabapathy K, Trp53-dependent DNA repair is affected by the codon 72 polymorphismOncogene 2006 25:3489-500.10.1038/sj.onc.120940516462765 [Google Scholar] [CrossRef] [PubMed]
[16]. Faghani M, Nikbahkt M, Salehi M, Rabbani M, Talebi A, Soleimani B, Study of p53 polymorphism at codon 72 in patients of breast cancer in IsfahanJ Isfahan Med School 2007 25:679-85. [Google Scholar]
[17]. Soleimani A, Rahmani Y, Farshchian N, Delpisheh A, Khassi K, Shahmohammadi A, The evaluation of p53 polymorphism at codon 72 and association with breast cancer in iran: a systematic review and meta-analysisJ Cancer Prev 2016 21(4):288-93.10.15430/JCP.2016.21.4.28828053964 [Google Scholar] [CrossRef] [PubMed]
[18]. Hattori Y, Ikeda Y, Suzuki Y, Ichikawa D, Matsushita M, Codon 72 polymorphism of TP53 gene is a novel prognostic marker for therapy in multiple myelomaBr J Haematol 2014 165:728-31.10.1111/bjh.1278424611901 [Google Scholar] [CrossRef] [PubMed]
[19]. Green MR, Sambrook J, Molecular cloning: A laboratory manual 2014 vol. 14th edNew YorkCold Spring Harbor Laboratory Press:47-53.:94-98. [Google Scholar]
[20]. Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics dataHum Mutat 2016 37(9):865-76.10.1002/humu.2303527328919 [Google Scholar] [CrossRef] [PubMed]
[21]. Pietsch EC, Humbey O, Murphy ME, Polymorphisms in the p53 pathwayOncogene 2006 25:1602-11.10.1038/sj.onc.120936716550160 [Google Scholar] [CrossRef] [PubMed]
[22]. Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, Genetics and cytogenetics of multiple myeloma: a workshop reportCancer Res 2004 64:1546-58.10.1158/0008-5472.CAN-03-287614989251 [Google Scholar] [CrossRef] [PubMed]
[23]. Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, Clinical significance of TP53 mutation in myelomaLeukemia 2007 21:582-84.10.1038/sj.leu.240452417215851 [Google Scholar] [CrossRef] [PubMed]
[24]. Lodé L, Eveillard M, Trichet V, Soussi T, Wuillème S, Richebourg S, Mutations in TP53 are exclusively associated with del(17p) in multiple myelomaHaematologica 2010 95(11):1973-6.10.3324/haematol.2010.02369720634494 [Google Scholar] [CrossRef] [PubMed]
[25]. Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D, TP53 Mutation and Survival in Chronic Lymphocytic LeukemiaJ Clin Oncol 2010 28:4473-79.10.1200/JCO.2009.27.876220697090 [Google Scholar] [CrossRef] [PubMed]
[26]. Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, Tseng MH, TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolutionBlood Cancer J 2015 5:e33110.1038/bcj.2015.5926230955 [Google Scholar] [CrossRef] [PubMed]
[27]. Kadia TM, Jain P, Ravandi F, Garcia-Manero G, Andreef M, Takahashi K, TP53 mutations in newly diagnosed acute myeloid leukemia: clinicomolecular characteristics, response to therapy, and outcomesCancer 2016 122:3484-91.10.1002/cncr.3020327463065 [Google Scholar] [CrossRef] [PubMed]
[28]. Takahashi Y, Ishii Y, Nagata T, Ikarashi M, Ishikawa K, Asai S, Clinical application of oligonucleotide probe array for full-length gene sequencing of TP53 in colon cancerOncology 2003 64:54-60.10.1159/00006651012457032 [Google Scholar] [CrossRef] [PubMed]
[29]. Yang Y, Wei Y, Bing-ji W, p53 mutations and protein overexpression in primary colorectal cancer and its liver metastasisChin J Cancer Res 2000 12(1):50-53.10.1007/BF02983194 [Google Scholar] [CrossRef]
[30]. Phatak P, Kalaisevi S, Divya T, Hegde AS, Hegde S, Kumaravel S, Alterations in tumour suppressor gene p53 in human gliomas from Indian patientsJ Biosci 2002 27(7):673-78.10.1007/BF0270837512571372 [Google Scholar] [CrossRef] [PubMed]
[31]. Berggren P, Hemminki K, Steineck G, the Stockholm Bladder Cancer Group. p53 intron 7 polymorphisms in urinary bladder cancer patients and controlsMutagenesis 2000 15(1):57-60.10.1093/mutage/15.1.5710640531 [Google Scholar] [CrossRef] [PubMed]
[32]. Prosser J, Condie A, Biallelic ApaI polymorphism of the human p53 gene (TP53)Nucleic Acids Res 1991 19:479910.1093/nar/19.17.4799-a1679931 [Google Scholar] [CrossRef] [PubMed]
[33]. Palmero EI, Alemar B, Schüler-Faccini L, Hainaut P, Moreira-Filho AC, Ewald IP, Screening for germline BRCA1, BRCA2, TP53 and CHEK2 mutations in families at-risk for hereditary breast cancer identified in a population-based study from Southern BrazilGenet Mol Biol 2016 39(2):210-22.10.1590/1678-4685-gmb-2014-036327223485 [Google Scholar] [CrossRef] [PubMed]
[34]. Djansugurova L, Zhunussova G, Khussainova E, Iksan O, Afonin G, Khaidarova D, Screening the APC, MLH1, MSH2 and TP53 mutations in patients with early onset of colorectal cancerJ Carcinog Mutagen 2014 5(6):19710.4172/2157-2518.1000197 [Google Scholar] [CrossRef]
[35]. Zhunussova G, Djansugurova L, Khussainova E, Zhunusbekova B, Afonin G, Khaidarova D, K-ras codon 12 and not TP53 mutations are predominant in advanced colorectal cancersS Afr Med J 2015 105(8):670-74.10.7196/SAMJnew.788626449693 [Google Scholar] [CrossRef] [PubMed]
[36]. Bansal A, Das P, Kannan S, Mahantshetty U, Mulherkar R, Effect of p53 codon 72 polymorphism on the survival outcome in advanced stage cervical cancer patients in IndiaInd J Med Res 2016 144(3):359-65.10.4103/0971-5916.19868528139534 [Google Scholar] [CrossRef] [PubMed]
[37]. Rao AKDM, Manikandan M, Arunkumar G, Revathidevi S, Vinothkumar V, Arun K, Prevalence of p53 codon 72, p73 G4C14-A4T14 and MDM2 T309G polymorphisms and its association with the risk of oral cancer in South IndiansGene Reports 2017 7:106-12.10.1016/j.genrep.2017.03.003 [Google Scholar] [CrossRef]
[38]. Ortega MM, Honma HN, Zambon L, Lorand-Metze I, Costa FF, De Souza CA, GSTM1 and codon 72 P53 polymorphism in multiple myelomaAnn Haematol 2007 86(11):815-19.10.1007/s00277-007-0347-x17653713 [Google Scholar] [CrossRef] [PubMed]