Introduction
Multidrug-Resistant Tuberculosis (MDR-TB) is a growing concern which has always played a role in controlling infectious diseases. Pyrazinamide (PZA) is one of the four front line cures of tuberculosis during the first two months of the treatment course. Diagnosis of PZA resistance is imperative in order to optimize the efficacy of new treatment regimens and minimize the risk of developing resistance to new drugs. In addition, high prevalence of PZA resistance, especially among those afflicted with MDR-TB, points to the need for developing those medicinal regimens that can be used in patients with PZA resistant MDR-TB.
Aim
The aim of this study was to determine anti-tuberculosis drug resistance rate and identify the correlation between MDR-TB and of mutations in pncA gene among Mycobacteriumtuberculosis isolates and frequency of mutations associated with PZA resistance in all the isolates.
Materials and Methods
This is a cross-sectional descriptive study conducted from April 2014 to June 2015. A total of 118 Mycobacterium tuberculosis (M. tuberculosis) clinical strains were isolates from patients referred to TB reference laboratory of Kermanshah from 8 provinces including: Kermanshah, Lorestan, Hamadan, Ilam, Kurdistan, Ardabil, Uromia and Tabriz. Antimicrobial susceptibility testing was performed using the proportional method and Minimum Inhibitory Concentration (MIC) and mutations in pncA for the PZA resistant isolates were studied using monoplex- Polymerase Chain Reaction (PCR) and then PCR products were sent for sequencing.
Results
Among the 118 clinical samples of M. tuberculosis investigated in various parts of Iran, 10 isolates (8.5%) were resistant to Isoniazid, 10 isolates (8.5%) were resistant to Rifampin, 7 isolates (6%) were MDR, and 23 isolates (19.5%) were resistant to PZA. Only did one MDR-TB isolate resistant (14.3%) to PZA show inactive mutation at Glu-122 codon that was found in pncA gene. According to our results, a significant correlation was found between MDR strains and of mutations in pncA gene (pv=0.049). pncA gene was not isolated from any of the PZA resistant isolates.
Conclusion
Our findings indicated that only one MDR-TB isolate resistant to PZA showed a mutation in the pncA gene (14.3%) and mutations were not observed in the other PZA-resistance isolates. The reason for resistance to PZA in the other PZA-resistance strains might be related to mutations in other genes or to some other factors. Thus, these reasons need to be further investigated in our study population.
Introduction
Despite massive efforts, Tuberculosis (TB) is one of the most important diseases that affect humans. In 2015, the World Health Organization (WHO) estimated that one-third of the world’s population was infected and reported an outbreak afflicting 9 million people with the total mortality of 1.7 million of them [1]. The rise and spread of drug-resistant strains of M. tuberculosis, in particular MDR strains, are crucial threats to the control of tuberculosis and constitute an important public health issue. Patients with MDR strains, as a species resistant to both Rifampin (RIF) and Isoniazid (INH), are hard to treat and more presumably stay as sources of infection for a longer duration than do patients with drug-sensitive strains [1-3]. Pyrazinamide (PZA) is the main first-line anti-tuberculosis (anti-TB) drug that is applied in short-period chemotherapy and is one of the main drugs in curing MDR [4]. PZA seems to destroy at least 95% of the half-persistent bacterial number persisting in a low pH setting because its activity is present only in the acidic environment found in active inflammation [5,6]. PncA gene encodes the pyrazinamidase enzyme which is an essential step to the activation of PZA. A mutation in the pncA gene leads to a decrease in the function of PZase enzyme which is the main resisting mechanism to Pyrazinamide. PZA needs enzyme transformation into its active type, Pyrazinoic acid, by the bacterial Pyrazinamidase (PZase), which is encoded by the 561- nucleotide (nt) pncA gene [7,8]. Mutations in the pncA end can stop or reduce PZase activity, which needs to be taken into account as the initial mechanism of PZA resistance in M. tuberculosis [9]. However, the diversity level of pncA mutations as it is described at present can be served as a marker when tracing the outbreak or transmission of PZA-resistant M. tuberculosis isolates. Interpretation of pncA mutation results for epidemiologic purposes needs to be cautiously done [10]. Specifying the frequency of mutations in resistant isolates of M. tuberculosis to PZA is significant in presenting proper treatment of tuberculosis in order to avoid the spread of infection in the society. Thus, the present study was to determine anti-tuberculosis drug resistant rate and identify the correlation between MDR-TB and of mutations in pncA gene among 118 M. tuberculosis strains and frequency of mutations associated with PZA-resistance in the M. tuberculosis Isolates who were referred to the reference TB laboratory of Kermanshah, west of Iran.
Materials and Methods
Mycobacterial Isolates
In the present study 135 smear-positive sputum samples were collected from patients suspected of TB from 8 provinces including Kermanshah, Lorestan, Hamadan, Ilam, Kurdistan, Ardabil, Uromia and Tabriz who were referred to the reference center in Kermanshah, from April 2014 to June 2015. Among these 135 samples, only 118 cases had positive culture (Positive culture refers to cases in which at least one colony was grown on Lowenstein-Jensen medium) in LJ medium and thus only these cases were included in the present study. Cases irrespective of age and sex were included. Samples that were culture negative were excluded from the present study. Colony form, growth rate and presence or absence of pigment was recorded and these 118 samples were identified as M. tuberculosis based on standard biochemical tests. Among these 118 patients 39 patients were under the age of 50, and 79 of them were aged above 50.
Bacterial culture was accomplished in the Lowenstein-Jensen (LJ) medium at 370C for 3 to 4 weeks and the isolates were identified by Ziehl-Neelsen method after decontamination by 4% NaOH [11] and biochemical tests such as: niacin production and nitrate and catalase tests [3]. Antimicrobial Susceptibility Testing (AST) was performed using the proportional method following the current recommendations from the World Health Organization (WHO) [12]. The critical concentrations were 0.2 mg/l for INH, 40mg/l for RIF, 2mg/l for Ethambutol (EMB), and 100 mg/l for PZA [13]. Determination of Minimum Inhibitory Concentration (MIC) for PZA in sputum preparation by the Broth Microdilution Method with 7H9 Broth [14]. Freshly grown colonies from LJ medium were transferred to a tube containing phosphate buffer saline to equal the density of 0.5 McFarland standard for use. All isolates were cultured in Middle brook 7H9 supplemented with Albumin-Dextrose-Catalase (ADC) at pH 5.5 containing PZA concentrations ranging from 100 to 800 mg/l. The PZA-susceptible M. tuberculosis strain H37Rv (ATCC 27294) was used as control. DNA extraction and PCR amplification chromosomal DNA were extracted from M. tuberculosis strains using the Qiagen kit according to manufacturer’s instructions (Qiagen GmbH, Hilden, Germany). A 670-bp segment of the pncA gene was amplified by using the conditions and the set of primers PF5 -bof 35 cycles of 95°C for 30 seconds for denaturation and 57°C for 30 seconds for annealing and 72°C for 45 seconds [15] elongation was performed with a BioRad thermal cycler PCR C1000 [11]. The expected size of the pncA PCR products was about 670 bp [16]. After electrophoresis of PCR products on 1% Agarose gel (Merck Co, Germany) and staining with Ethidium bromide, the DNA bands were visualized by Gel documentation apparatus (BioRad, USA). PCR, sequencing and bioinformatic analyses were performed to identify mutations in pncA genes. To determine the pncA sequence, PCR products was sent for sequencing. The sequencing was performed using Dye terminator sequencing method (ABI 3730XL DNA analyser apparatus Macrogen Inc., Korea). Sequenced data were edited by using BioEdit software version 7.05.3, and the results were compared with the H37Rv genome. The GenBank accession number for pncA gene in this work was KY659393 [17].
Statistical Analysis
All data were analysed by using SPSS version 23.0 for the correlation between gene mutation and the resistance by Chi-square test. Statistical significance was defined as p-value ≤ 0.05.
Ethics
The project was approved by the Ethics Committee and we have received code of Ethics with number IR.KUMS.REC.1394.504.
Results
A total of 118 M. tuberculosis clinical strains were exclusively isolated from sputum samples collected from patients who were diagnosed with susceptible or resistant TB reference laboratory of Kermanshah for MDR-TB cases [Table/Fig-1]. 69 (58.4%) of them were male and 49 (41.5%) were female. The average age of patients was 47 years. While 117 of them were newly infected, 1 of them was reinfected. Among 118 studied patients, 10 samples (8.5%) were resistant against isoniazid, 10 samples (8.5%) were resistant against Rifampin, 7 samples (6%) were MDR, and 23 (19.5%) were resistant to PZA [Table/Fig-2]. MIC for four different concentrations is shown in [Table/Fig-3]. Analysis of mutations associated with PZA resistance isolates [Table/Fig-4] showed that only in one isolate was an one-point mutation for codon of Glu-122 in (GAG to GAA) the pncA gene. The GenBank accession number for pncA gene in this work was KY659393. According to our results, there was no significant relation between resistance to PZA and mutations in pncA gene (p-value ≥0.05) and a significant correlation between was found MDR strains and of mutations in pncA gene (p=0.049). pncA gene was not isolated from any of the PZA resistant isolates.
Prevalence of MTB drug resistance in this study.
| Province | Positive sample of MTB | Rifampin resistance | Isoniazid resistance | Pyrazinamide resistance |
|---|
| Ardabil | 9(7.62%) | 0 | 1(10%) | 1(4.34%) |
| Tabriz | 21(17.7%) | 2(20%) | 2(20%) | 6(26%) |
| Urmia | 10(8.47%) | 0 | 0 | 3(13.04%) |
| Ilam | 4(3.38%) | 0 | 0 | 1(4.34%) |
| Kurdistan | 27(22.8%) | 0 | 0 | 3(13.04%) |
| Kermanshah | 17(14.4%) | 2(20%) | 1(10%) | 2(8.69%) |
| Lorestan | 19(16.1%) | 0 | 0 | 4(17.39%) |
| Hamadan | 5(4.2%) | 0 | 0 | 2(8.69%) |
| Others | 6(5%) | 6(60%) | 6(60%) | 1(4.34%) |
| Total | 118 | 10 | 10 | 23 |
Results in proportional method.
| Anti-microbial drug | Resistance; No. (%) | Sensitive; No. (%) | Total |
|---|
| Isoniazid | 10(8.5%) | 108(91.5%) | 118(100%) |
| Rifampin | 10(8.5%) | 108(91.5%) | 118(100%) |
| Pyrazinamide | 23(19.5%) | 95(80.5%) | 118(100%) |
| MDR | 7(6%) | 111(94%) | 118(100%) |
MIC of Pyrazinamide resistant.
| MIC & Anti- microbial Test | 100 μg/ml | 200 μg/ml | 400 μg/ml | >800 μg/ml |
|---|
| Pyrazinamide | 9(39.1%) | 3(13%) | 4(17.39%) | 7(30%) |
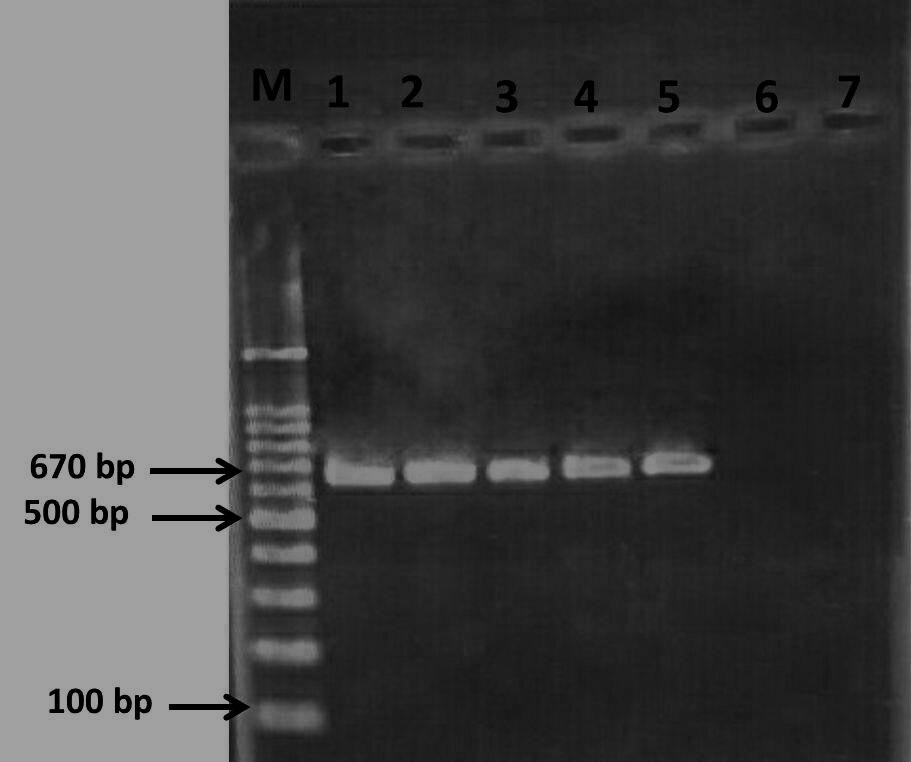
Polymerase chain reaction image of the PCR products (pncA gene) on a 1.2% agaros gel. Of right to left: Lane M = 100-bp ladder DNA size marker; lane 1-5 = M. tuberculosis clinical isolates (positive sample for pncA gene); lane 6 and 7, negative control.
Discussion
Tuberculosis, which is caused by M. tuberculosis, has long been recognized as a major global calamity. It is recognized as the eighth cause of death worldwide and the second most prevalent fatal infectious disease [18]. Although different kinds of medications have been used to cure tuberculosis for many years, it has continued to remain as a major universal concern. The advent of drug-resistant strains and genotypes has posed serious hurdles in the path to control and treat this disease. Some strains of the bacterium have drawn more attention for the same reason [19]. Multi-drug resistance of various strains of M. tuberculosis has turned into one of the most serious challenges in the treatment of tuberculosis [20,21]. Zhang H et al., reported a multiple-drug resistance of 77% in China [22]. In a systematic review study in Iran, the findings of Nasiri MJ et al., showed that 23 percent of new cases of tuberculosis and 65.6 percent of previous cases of the disease treated in the past were resistant to at least one the anti-TB medications [23]. Our findings suggest that out of the 118 cases of the study, 7 cases (6%) of the isolates possessed multi-drug resistance. Similar studies reported the following percentages of 7 multi-drug resistance among the isolates: 3.4% in Golestan, 2.9% in Tabriz and 15% in Tehran Province [24-26]. PZA is considered as one of the four first line drugs for curing TB during the first two months of the treatment course [27]. Today, a high prevalence of resistance to PZA has been reported worldwide. Various studies have deduced that different mechanisms play a part in the development of PZA resistance [28]. In our study, 23 isolates (19.5%) out of 118 samples were PZA resistant, whereas the findings of Xia Q et al., in China, Maslov DA et al., in Russia and Jonmalung J et al., in Thiland indicated that 43.7%, 74.3% and 49% of the isolates were PZA resistant, respectively [15,29,30]. However, Sreevatsan S et al., pointed out that 72% of the PZA resistant isolates in their study possessed a mutation in the pncA gene [31]. In addition, the findings of Cuevas-Córdoba B et al., in Mexico, Jnawali HN et al., in Korea, Zhao L-I et al., in China, and Perdigao J et al., in Portugal showed 81%, 87.8%, 11.5%, and 25% of the PZA resistant isolates revealed mutations in the pncA gene, respectively [32-35]. According to our findings, out of the 23 PZA resistant cases and 7 MDR isolates, a mutation in pncA gene was observed only in one case (14.3%). The results of Doustdar F et al., study on M. tuberculosis isolates resistant to PZA in Tehran showed that in 8 PZA resistant isolates with negative PZase activity, there was no mutation in the pncA gene [28]. In a study by Akhmetova A et al., in Kazakhstan, out of 36 pyrazinamide-sensitive cases (46.7%), two mutations (5.5%) in the pncA were detected [36]. The results of Li H et al., in China also showed that there were mutations in the pncA gene in three pyrazinamide-sensitive mycobacterium cases [37]. Sreevatsan S et al., indicated that in 28% of pyrazinamide-resistant isolates there was no mutation in the pncA gene [31]. However, the results of other studies have also shown that the rpsA gene, which encodes the S1 ribosomal protein and is a vital protein involved in the transfer of protein and ribosome secretion, can cause pyrazinamide resistance as well. Alanine deletion in the C-terminal of rpsA gene leads to dramatic increase in PZA resistance [38]. In a similar study by Xia Q et al., on 118 pyrazinamide-resistant mycobacteria and 161 sensitive cases, 92 resistant and 5 sensitive cases in the pncA gene and also 5 pyrazinamide-resistant and 6 sensitive cases in the rpsA gene exhibited mutations [15]. Cui Z et al., studied 423 cases of M. tuberculosis in China and found that in four pyrazinamide-resistant cases with no change in the pncA gene, there was deleting mutation in the rpsA gene [39]. The findings of Zhang S et al., showed that out of 174 pyrazinamide-resistant mycobacterium cases, in 5(3%) of them no mutations in the pncA and rpsA genes were observed. It also revealed that mutation in the panD gene was closely related to pyrazinamide resistance. This gene is encoded by the aspartate decarboxylase enzyme [40]. The results of sequencing 30 cases of pyrazinamide-resistant mycobacterium by Shi W et al., disclosed no mutations in the pncA and rpsA genes while 24 cases (80%) revealed mutations in the panD gene [38]. Two more mechanisms recently proposed for pyrazinamide resistance include efflux pumps and a flaw in the absorption of pyrazinamide by the organism [41]. Zimic M et al., showed that changes in the amount of bacterial Pyrazinoic acid (POA) exit (POA output) affects PZA resistance. These changes might depend on the levels of PZAse activity, PZAse intracellular concentration, and the efficiency of POA exit pump [41]. Although different results have indicated that PZA resistant mutation in the pncA gene are not only highly variable and disperse throughout the gene but various levels of PZA resistance might be observed. The results of our study and similar ones in Iran, however, showed that the frequency of mutations related to PZA resistance in the pncA gene among M. tuberculosis cases is very rare in the west of Iran. As mentioned, numerous mechanisms play a role in pyrazinamide resistance. Given the widespread development of M. tuberculosis and highly dynamic mutations in the pncA gene related to PZA resistance, it is expected that newer mutations in the pncA gene and the effects of other genes on pyrazinamide resistance among clinical cases of M. tuberculosis in the west of Iran be observed in the future.
Limitation
Our study has several limitations; one limitation of our study is that a small number of MDR isolates were observed in our samples. Therefore, the possibility of resistance to PZA and the presence of mutation in the pncA gene has also been low, leading to a decrease in the prevalence of resistance to PZA due to mutations in the pncA gene in our study. Another limitation of our study concerns investigating mutations only in the pncA gene, ignoring resistance caused by mutations in the other genes if mutations in the other genes had also been studied. Despite these limitations, our study is important because it demonstrates the problem of resistance to PZA in eight provinces of Iran and treated for PZA-resistance M. tuberculosis strains.
Conclusion
Our findings indicated that only one MDR-TB isolate resistant to PZA showed a mutation in the pncA gene (14.3%) and mutations were not observed in the other PZA-resistance isolates. The reason for resistant to PZA in the other PZA-resistance strains might be related to mutations in other genes or to some other factors in our study population. Therefore, there is a higher possibility for mutations to occur in the MDR strains of M. tuberculosis. Our results illustrate the need for further research to investigate a more resistant gene association with PZA resistance in PZA-resistance M. tuberculosis strains.
[1]. Mohajeri P, Moradi S, Atashi S, Farahani A, Mycobacterium tuberculosis Beijing Genotype in Western Iran: Distribution and Drug ResistanceJournal of Clinical and Diagnostic Research: JCDR 2016 10(10):DC05 [Google Scholar]
[2]. Gu Y, Yu X, Jiang G, Wang X, Ma Y, Li Y, Pyrazinamide resistance among multidrug-resistant tuberculosis clinical isolates in a national referral center of China and its correlations with pncA, rpsA, and panD gene mutationsDiagnostic microbiology and infectious disease 2016 84(3):207-11. [Google Scholar]
[3]. Mohajeri P, Sadri H, Farahani A, Norozi B, Atashi S, Frequency of mutations associated with rifampicin resistance in Mycobacterium tuberculosis strains isolated from patients in West of IranMicrobial Drug Resistance 2015 21(3):315-19. [Google Scholar]
[4]. Hoffner S, Ängeby K, Sturegård E, Jönsson B, Johansson A, Sellin M, Proficiency of drug susceptibility testing of Mycobacterium tuberculosis against pyrazinamide: the Swedish experienceThe International Journal of Tuberculosis and Lung Disease 2013 17(11):1486-90. [Google Scholar]
[5]. Piersimoni C, Mustazzolu A, Giannoni F, Bornigia S, Gherardi G, Fattorini L, Prevention of false resistance results obtained in testing the susceptibility of Mycobacterium tuberculosis to pyrazinamide with the Bactec MGIT 960 system using a reduced inoculumJournal of Clinical Microbiology 2013 51(1):291-94. [Google Scholar]
[6]. Stoffels K, Mathys V, Fauville-Dufaux M, Wintjens R, Bifani P, Systematic analysis of pyrazinamide-resistant spontaneous mutants and clinical isolates of Mycobacterium tuberculosisAntimicrobial Agents And Chemotherapy 2012 56(10):5186-93. [Google Scholar]
[7]. Shi W, Chen J, Feng J, Cui P, Zhang S, Weng X, Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosisEmerg Microbes Infect 2014 3:e58CrossRef PubMed PubMedCentral Google Scholar. 2014 [Google Scholar]
[8]. Sharma B, Pal N, Malhotra B, Vyas L, Rishi S, Comparison of MGIT 960 & pyrazinamidase activity assay for pyrazinamide susceptibility testing of Mycobacterium tuberculosisIndian J Med Res 2010 132:72-6. [Google Scholar]
[9]. Perdigão J, Macedo R, Malaquias A, Ferreira A, Brum L, Portugal I, Genetic analysis of extensively drug-resistant Mycobacterium tuberculosis strains in Lisbon, PortugalJournal of Antimicrobial Chemotherapy 2009 65(2):224-27. [Google Scholar]
[10]. Napiórkowska A, Rüsch-Gerdes S, Hillemann D, Richter E, Augustynowicz-Kopeć E, Characterisation of pyrazinamide-resistant Mycobacterium tuberculosis strains isolated in Poland and GermanyThe International Journal of Tuberculosis and Lung Disease 2014 18(4):454-60. [Google Scholar]
[11]. Muthaiah M, Jagadeesan S, Ayalusamy N, Sreenivasan M, Prabhu SS, Muthuraj U, Molecular epidemiological study of pyrazinamide-resistance in clinical isolates of Mycobacterium tuberculosis from South IndiaInternational Journal Of Molecular Sciences 2010 11(7):2670-80. [Google Scholar]
[12]. World Health Organization, 2009. WHO/HTM/TB/2009.411. Geneva, Switzerland: WHO; 2009. Global Tuberculosis Control: Epidemiology, Strategy, Financing: WHO Report 2009 [Google Scholar]
[13]. Mohajeri P, Norozi B, Atashi S, Farahani A, Anti tuberculosis drug resistance in west of IranJournal of Global Infectious Diseases 2014 6(3):114 [Google Scholar]
[14]. Campanerut PAZ, Ghiraldi LD, Spositto FLE, Sato DN, Leite CQF, Hiroyuki Hirata M, Rapid detection of resistance to pyrazinamide in Mycobacterium tuberculosis using the resazurin microtitre assayJournal of Antimicrobial Chemotherapy 2011 66(5):1044-46. [Google Scholar]
[15]. Xia Q, Zhao L-l, Li F, Fan Y-m, Chen Y-y, Wu B-b, Phenotypic and genotypic characterization of pyrazinamide resistance among multidrug-resistant Mycobacterium tuberculosis isolates in Zhejiang, ChinaAntimicrobial Agents and Chemotherapy 2015 59(3):1690-95. [Google Scholar]
[16]. Sekiguchi J-I, Nakamura T, Miyoshi-Akiyama T, Kirikae F, Kobayashi I, Augustynowicz-Kopeć E, Development and evaluation of a line probe assay for rapid identification of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis strainsJournal of Clinical Microbiology 2007 45(9):2802-07. [Google Scholar]
[17]. Moradi J, Mohajeri P, Alvandi A, Farahani A, Atashi S, Mycobacterium tuberculosis strain T38 pncA (pncA) gene, complete cdsAvailable from: https://www.ncbi.nlm.nih.gov/nuccore/KY659393 [Google Scholar]
[18]. Wengenack NL, Lane BD, Hill PJ, Uhl JR, Lukat-Rodgers GS, Hall L, Purification and characterization of Mycobacterium tuberculosis KatG, KatG (S315T), and Mycobacterium bovis KatG (R463L)Protein expression and purification 2004 36(2):232-43. [Google Scholar]
[19]. Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, ZiaZarifi AH, Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in IranChest Journal 2009 136(2):420-25. [Google Scholar]
[20]. Sadri H, Farahani A, Mohajeri P, Frequency of mutations associated with isoniazid-resistant in clinical Mycobacterium tuberculosis strains by low-cost and density (LCD) DNA microarraysAnnals of Tropical Medicine and Public Health 2016 9(5):307-11. [Google Scholar]
[21]. Mohajeri P, Norozi B, Atashi S, Farahani A, Anti tuberculosis drug resistance in west of iranJ Glob Infect Dis 2014 6(3):114-17. [Google Scholar]
[22]. Zhang H, Bi L, Li C, Sun Z, Deng J, Zhang X, Mutations found in the pncA gene of Mycobacterium tuberculosis in clinical pyrazinamide-resistant isolates from a local region of ChinaJournal of International Medical Research 2009 37(5):1430-35. [Google Scholar]
[23]. Nasiri MJ, Dabiri H, Darban-Sarokhalil D, Rezadehbashi M, Zamani S, Prevalence of drug-resistant tuberculosis in Iran: systematic review and meta-analysisAmerican Journal of Infection Control 2014 42(11):1212-18. [Google Scholar]
[24]. Livani S, Mirinargesi M, Nemati-Shoja E, Rafiei S, Taziki M, Tabarraei A, Prevalence ofMultidrug Resistant Mycobacterium tuberculosis by Mycobacteria growth indicator tube in Golestan province, North of IranMedical Laboratory Journal 2011 5(2):7-14. [Google Scholar]
[25]. Roshdi Maleki M, Moaddab S, Drug susceptibility pattern of Mycobacterium tuberculosis strains to first and second line drugs in Tabriz, IranIranian Journal of Medical Microbiology 2009 3(1):18-24. [Google Scholar]
[26]. Farnia P, Masjedi MR, Mirsaeidi M, Mohammadi F, Vincent V, Bahadori M, Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patientsJournal of Infection 2006 53(5):331-36. [Google Scholar]
[27]. Zhang Y, Shi W, Zhang W, Mitchison D, Mechanisms of pyrazinamide action and resistanceMicrobiol Spectr 2013 2(4):1-12.MGM2-0023-2013 [Google Scholar]
[28]. Doustdar F, Khosravi AD, Farnia P, Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of IranMicrobial Drug Resistance 2009 15(4):251-56. [Google Scholar]
[29]. Maslov DA, Zaĭchikova MV, Chernousova LN, Shur KV, Bekker OB, Smirnova TG, Resistance to pyrazinamide in Russian Mycobacterium tuberculosis isolates: pncA sequencing versus Bactec MGIT 960Tuberculosis 2015 95(5):608-12. [Google Scholar]
[30]. Jonmalung J, Prammananan T, Leechawengwongs M, Chaiprasert A, Surveillance of pyrazinamide susceptibility among multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, ThailandBMC microbiology 2010 10(1):223 [Google Scholar]
[31]. Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM, Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organismsAntimicrobial agents and chemotherapy 1997 41(3):636-40. [Google Scholar]
[32]. Cuevas-Córdoba B, Xochihua-González SO, Cuellar A, Fuentes-Domínguez J, Zenteno-Cuevas R, Characterization of pncA gene mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from MexicoInfection, Genetics and Evolution 2013 19:330-34. [Google Scholar]
[33]. Jnawali HN, Hwang SC, Park YK, Kim H, Lee YS, Chung GT, Characterization of mutations in multi-and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in Republic of KoreaDiagnostic microbiology and infectious disease 2013 76(2):187-96. [Google Scholar]
[34]. Zhao L-l, Chen Y, Chen Z-n, Liu H-c, Hu P-l, Sun Q, Prevalence and molecular characteristics of drug-resistant Mycobacterium tuberculosis in Hunan, ChinaAntimicrobial agents and chemotherapy 2014 58(6):3475-80. [Google Scholar]
[35]. Perdigao J, Macedo R, Joao I, Fernandes E, Brum L, Portugal I, Multidrug-resistant tuberculosis in Lisbon, Portugal: a molecular epidemiological perspectiveMicrobial Drug Resistance 2008 14(2):133-43. [Google Scholar]
[36]. Akhmetova A, Kozhamkulov U, Bismilda V, Chingissova L, Abildaev T, Dymova M, Mutations in the pncA and rpsA genes among 77 Mycobacterium tuberculosis isolates in KazakhstanThe International Journal of Tuberculosis and Lung Disease 2015 19(2):179-84. [Google Scholar]
[37]. Li H, Chen J, Zhou M, Geng X, Yu J, Wang W, Rapid detection of Mycobacterium tuberculosis and pyrazinamide susceptibility related to pncA mutations in sputum specimens through an integrated gene-to-protein function approachJournal of Clinical Microbiology 2014 52(1):260-67. [Google Scholar]
[38]. Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosisScience 2011 333(6049):1630-32. [Google Scholar]
[39]. Cui Z, Wang J, Lu J, Huang X, Zheng R, Hu Z, Evaluation of methods for testing the susceptibility of clinical Mycobacterium tuberculosis isolates to pyrazinamideJournal of Clinical Microbiology 2013 51(5):1374-80. [Google Scholar]
[40]. Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y, Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosisEmerging Microbes & Infections 2013 2(6):e34 [Google Scholar]
[41]. Zimic M, Fuentes P, Gilman RH, Gutiérrez AH, Kirwan D, Sheen P, Pyrazinoic acid efflux rate in Mycobacterium tuberculosis is a better proxy of pyrazinamide resistanceTuberculosis 2012 92(1):84-91. [Google Scholar]