Perinatal mortality is a major challenge for health services in the developing world and FGR has long been accepted as one of the leading causes [1]. Although the occurrence of FGR is seen in the developed world as well, the rate of incidence (about 10%) [2] is quite alarming in the developing world and India accounts for 43.6% of FGR among developing nations [3]. The ill effects of FGR are not limited to the perinatal period and infancy, rather, the sequelae persists in adulthood and predisposes the individual to increased incidence of several non-communicable disorders in later stages of life, including cardiovascular disease, diabetes mellitus and metabolic syndrome [4-7].
Various factors predispose to FGR, which can be categorised based on their origin. Maternal morbidity, addictions and nutritional status (both before and during pregnancy) affect the foetal development in utero. Foetal structural and chromosomal anomalies, infections and multiple pregnancies have adverse effect on intrauterine growth. Placental abnormalities also restrict the growth of the foetus. However, about 40% of FGR are still categorised as ‘idiopathic’ [8].
Paraoxonase 1 (PON1) is a High Density Lipoprotein (HDL) bound enzyme, the major contributor to the anti-oxidant action of HDL that can regulate the levels of Oxidative Stress (OS) [9] through the reduction of oxidised Low Density Lipoprotein (LDL) [10]. Chen D et al., hypothesised that PON1 activity, which is well known for its antioxidant property towards LDL, when decreased, could be related to vascular endothelial damage, placental insufficiency and thrombosis and thus, could lead to adverse pregnancy outcomes [11].
Variation of paraoxonase activity among individuals related to PON1 gene polymorphism is quite prominent. Among the several polymorphic variants of the gene, the most commonly encountered types are the Q192R variant, the L55M variant (both in the coding regions) and the C108T variant (in the promoter region) [12]. Q192R polymorphism has been shown to be strongly correlated with variation in the paraoxonase activity. Moreover, Q192R genotype and paraoxonase activity have shown significance in the prediction of coronary artery disease [13]. No significant literature is available on association of PON1 polymorphism with IFGR, although studies with unfavourable pregnancy outcomes have been conducted [14]. The infant and maternal PON1-192R allele have been shown to be associated with preterm delivery in studies conducted in Chinese population and British gravida women [11,15].
Interestingly, a host of non-communicable disorders, which have been found to be associated with FGR, have recently been shown to also be associated with PON1 polymorphism. Diabetic nephropathy [16], metabolic syndrome [17] and coronary artery disease [18] are all more pronounced and comparatively more frequent in individuals with unfavourable genotype for PON1. This association has been found to persist across all regions in different ethnic groups (Caucasian and Asian) with strong statistical significance [16-18].
The biochemical data related to paraoxonase enzyme activity has already been published as part of assessment of oxidative stress in the same group of patients in relation to IFGR [19]. This study was designed to explore the relationship of foetal growth restriction with PON1 gene polymorphism and expression in IFGR neonates and their mothers.
Materials and Methods
The study was designed as a cross-sectional, case-control study and was conducted in the Department of Biochemistry in association with the Department of Obstetrics and Gynaecology, University College of Medical Sciences and Guru Teg Bahadur Hospital, New Delhi, from December 2011 to March 2013. Ethical clearance was obtained from the Institutional Ethical Committee Human Research. Informed written consent was taken from each subject before recruitment. In the present study, we analysed PON1 Q192R (rs662) polymorphism and estimated mRNA expression in whole blood and PON1 activity in the serum of IFGR neonates and their mothers, along with their controls.
Subjects
A total of 75 unrelated live births and their mothers were included (referred subsequently as IFGR neonates and IFGR mothers) with 75 matched controls.
For cases, pregnancies between 37-40 weeks with neonatal birth weight <10th percentile for that gestational age were selected. Mothers were in the age group of 18-35 years, had a pre-pregnancy Body Mass Index (BMI) of 19-26 kg/m2 and were singleton pregnancies. Exclusion criteria included; parity >4, chronic maternal diseases (including hypertension, diabetes mellitus, renal disease, liver disease, heart disease and moderate/severe anaemia), antepartum haemorrhage-abruptio placentae, placenta previa, pre-eclampsia, eclampsia, Anti Phospho-Lipid Antibody (APLA) syndrome and known parental genetic disease, history of substance abuse (smoking, alcohol and drugs), evidence of urinary tract infection or any intrauterine infection, gross placental abnormality, or any known cause of foetal growth restriction in history, examination and investigations. Cases with ultrasonologically detected anomalies in foetus were also excluded.
Information was collected and recorded for each enrolled subject for the mode of delivery, neonatal gestational age and anthropometry (birth weight, length and circumference of mid arm, head and chest). At the time of delivery, the attending obstetrician also carried out physical examination as well as measurement of weight of the placenta. Physical examination included inspection of maternal and foetal surfaces of placenta as well as the site of cord insertion for presence or absence of any calcifications, membrane, any obvious abnormality and any retroplacental clots. Presence or absence of all cotyledons and number of vessels in the cord were also examined. Any abnormality was ground for exclusion, as mentioned in exclusion criteria and deliveries with only normal appearing placenta were included in the study.
Women with low risk pregnancies between 37-40 weeks of gestation, with normal neonates based on gestational age and anthropometric measurements (birth weight, length and circumference of mid-arm, head and chest) were recruited as controls. Other inclusion and exclusion criteria were same as those for cases.
Haemoglobin level, blood group, VDRL test (Venereal Disease Research Laboratory), VCT (Voluntary Counseling and Testing for HIV), GCT (Glucose Challenge Test), urine (routine/microscopy and culture) and obstetrical ultrasound were carried out for all recruited subjects. Special investigations, including APLA (Antiphospho-lipid antibodies), TORCH (Toxoplasmosis, Rubella, Cytomegalo-virus, Herpes Simplex Virus-2, and other infections) and targeted obstetrical scan were carried out for subjects.
Sample Collection
A total of 5 mL each of maternal venous blood and cord blood was collected {2 mL in Ethylenediaminetetraacetic acid (EDTA) vacutainer and 3 mL in plain vacutainer}. For genomic study, 500 μL whole blood was kept separately at 4°-8°C and DNA was isolated within two weeks of collection. For mRNA isolation, 350 μL of whole blood was diluted with equal volume of Nuclease Free Water (NFW) and subsequently mixed with TRIzol LS (Invitrogen) in a ratio of 1:3 and stored at -80°C. RNA extraction was carried out within a month of sample collection. Serum was collected and kept at -80°C in aliquots for estimation of the different parameters till further use.
DNA Extraction, Quantification and Genotyping Analysis
Genomic DNA was extracted from whole blood by commercially available kit (Quick-g DNATM MiniPrep, Zymo Research) as per the instructions provided by the manufacturer. The extracted DNA was quantified by taking Optical Density (OD) of the sample at 260 nm by spectrophotometer (Thermo Scientific NanoDrop 2000c). The purity of DNA was ascertained by taking ratio of ODs at 260 nm and 280 nm.
PON1 Genotyping
PCR amplification was done using thermo cycler (Eppendorf Mastercycler Gradient-5331). Briefly, 50 ng of DNA was amplified in a 20 μL reaction mixture containing 15 pM of each of the PON1 primers, 1.25 mM MgCl2, 200 μM Deoxynucleotide Triphosphates (dNTPs), 2 μL 10X PCR buffer (10X buffer containing 500 mM KCl, 100 mM Tris-HCl, pH 9.0), and 1 U/μL Taq DNA polymerase (All reagents form MBI Fermentas). The PCR protocol included an initial incubation at temperature of 94°C (five minutes) followed by 35 cycles of amplification (denaturation for 30 seconds at 94°C, annealing for 30 seconds at 60°C and extension for 30 seconds at 72°C). A final 10 minutes extension step was done at 72°C. The primer sequences used are given in [Table/Fig-1].
Primer sequences for PON1 genotyping (rs662). The size before treatment was 99bp and size after treatment with RE 66bp and 33bp. Reference: Lakshmy R et al., [20].
| For PON1: | F: 5’-TATTGTTGCTGTGGGACCTGAG-3’ |
| R: 5’-CACGCTAAACCCAAATACATCTC-3’ |
The final PCR product from amplification of PON1 was subjected to digestion by BspPI Restriction Endonuclease (RE) enzyme (Thermo Scientific). A 10 μL of PCR reaction mixture was mixed with 2 U/μL of BspPI RE enzyme, 2 μL of 10X Buffer Tango and 18 μL of NFW. The mixture was incubated at 55°C for 16 hours. The enzyme was then inactivated by incubation at 80°C for 20 minutes. The digested product was visualised after electrophoresis in Ethidium Bromide (EtBr)-stained 2.5% agarose gel on gel documentation system (UVI-TEC).
Analysis of Gene Expression
Expression of the genes was assessed by RT-PCR/chemical assays. Only representative samples (15 samples) in each group, viz., cases and controls, were processed to establish correlation between PON1 mRNA expression and development of IFGR. Total RNA was extracted from peripheral blood kept in EDTA anticoagulant with TRIzol LS (Invitrogen, Carlsbad, CA). The extracted RNA was found to have an OD 280/260 ratio between 1.8 and 2.0. 500 ng of RNA was mixed with Oligo-dT and random hexamer (from Sigma Aldrich), and incubated at 65°C for five minutes followed by incubation on ice till addition of next set of reagents. The final mixture of 20 μL, which was used for synthesising cDNA, contained 4 μL of 5X Reaction Buffer (MBI Fermentas), 40 U/μL of Ribolock® (MBI Fermentas), 200 U/μL of RevertAid® (MBI Fermentas) and 200 μM each of dNTPs. The mixture was incubated at 25°C for 10 minutes, 45°C for 60 minutes, followed by 70°C for 10 minutes in a thermocycler (Eppendorf Mastercycler Gradient-5331). The diluted solution (1:4 dilution with NFW) was then used as template sample for qPCR. The reaction mixture contained 3 μL of template cDNA, 5 μL of Pyrostart Fast PCR Mix ((MBI Fermentas), 1 μL of Syto 9 dye (50 μM) and 15 pM each of forward and reverse primers [Table/Fig-2].
Primer sequences for PON1 qPCR. Reference: Parker-Katiraee L et al., [21].
| PON1: |
| Forward Primer: 5’-TATTGTTGCTGTGGGACCTGAG-3’ |
| Reverse Primer: 5’-CCACAGATATGTTATCCACG-3’. |
| GAPDH: |
| Forward Primer: 5’-CCAAGGTCATCCATGACAACTTTGGT-3’ |
| Reverse Primer: 5’-TGTTGAAGTCAGAGGAGACCACCTG-3’ |
The PCR protocol included an initial activation temperature of 95°C (one minute) followed by 35 cycles of amplification (15 seconds at 95°C, 15 seconds at 54.8°C and 30 seconds at 72°C). The fluorescence was acquired at 72°C. The results were analysed as described below. Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) gene was used as an internal control. Differences in threshold cycle (ΔCt) values for PON1 and housekeeping gene from qPCR were calculated for both cases and controls, which were then used to derive the difference of ΔCt between cases and controls (ΔΔCt) values for PON1 mRNA expression in foetus and mothers. Gene expression normalization (using GAPDH gene) was done by determining delta Ct, where ΔCt = average Ct (target) – average Ct (normaliser). Then the difference of mean Ct values between test and control was determined, ΔΔCt = ΔCt (control) – ΔCt (test). Determination of FC was based on FC = Primer Efficiency ΔΔCt. True fold change was represented by the following: if FC > 1, true fold change = FC; if FC < 1, true fold change = -1/FC.
Biochemical PON1 Activity
Paraoxonase enzyme activity was estimated following the conversion of paraoxon to p-nitro phenol. The 1 mL assay mixture contained 10 mM Tris HCl, 1 M NaCl and 2 mM CaCl2. 0.025 mL of serum was present in the assay mixture. Paraoxon was added at a concentration of 1.5 mM of final mixture. The rate of formation of p-nitro phenol was monitored by taking readings at every consecutive minute for five minutes at 405 nm using spectrophotometer (Shimadzu UV-2450). Extinction coefficient (17,000 M-1 cm-1) was used to calculate the enzyme activity and it was expressed as nmol min-1 mL-1 of serum [20].
Statistical Analysis
Statistical analysis was done by SPSS software version 20.0. For comparisons of demographic and biochemical data, student t-test and Chi-square test were used. The genotypes were compared using one-way ANOVA. Binary logistic was used to find out the Odds Ratio (OR) for IFGR amongst the different genotypes. The QQ genotype was considered as reference. A p-value < 0.05 was taken as statistically significant.
True FC was used to assess change in gene expression. Finally, the linear correlation of maternal and foetal PON1 with respect to foetal weight was estimated using SPSS software. One-way ANOVA with linear trend was applied to find the mean trend of maternal paraoxonase activity with RR, QR and QQ genotypes. Since no previous study has been done for PON1 in IFGR mother-neonate dyads, power of study for the selected sample size could not be determined.
Results
Gene Polymorphism
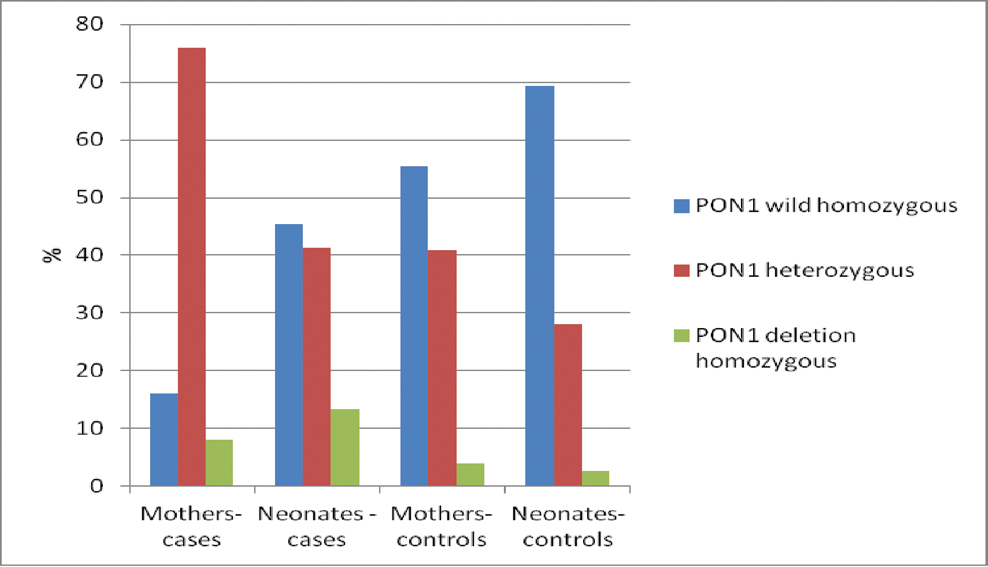
The prevalence of Q192R polymorphic variants of PON1 gene is shown in [Table/Fig-3] for the four study groups namely, mothers of IFGR neonates, control mothers, IFGR neonates and control neonates. In mothers of IFGR neonates, the frequency of RR and QR genotype as well as R allele was found to be significantly higher as compared to controls [Table/Fig-4a]. Presence of RR and QR genotype was found to be significantly associated with the incidence of IFGR (OR = 7, p-value=0.013 and OR=6.435, p-value <0.001 respectively).
Prevalence of PON1 Q192R genotypes in IFGR materno-foetal dyads with controls. The prevalence of various genotypic variants in each subject group is represented in the figure as percentages, where the bar represents the percentage of subjects in the depicted genotype.
Odds ratio of PON1 Q192R genotypes in mothers.
| Groups Genotypes | Mothers (cases) | Mothers (controls) | Odds Ratio | 95% CI | p-value |
|---|
| QQ | 12 (16%) | 41 (54.7%) | 1 | Ref | Ref |
| QR | 57 (76%) | 31 (41.3%) | 6.435 | 2.96-13.98 | <0.001* |
| RR | 6 (8%) | 3 (4%) | 7.00 | 1.52 – 32.33 | 0.013* |
| Allele Frequency |
| Q | 0.54 | 0.753 | - | - | - |
| R | 0.46 | 0.247 | - | - | - |
Similarly, in IFGR neonates, the frequency of RR and QR genotype as well as R allele was found to be significantly higher as compared to controls. Presence of RR and QR genotype was found to be significantly associated with the incidence of IFGR (OR=7.64, p-value=0.012 and OR=2.258, p-value=0.023 respectively) [Table/Fig-4b].
Odds ratio of PON1 Q192R genotypes in IFGR babies.
| Groups Genotypes | Neonates (cases) | Neonates (controls) | Odds Ratio | 95% CI | p-value |
|---|
| QQ | 34 (45.3%) | 52 (69.3%) | 1 | Ref | Ref |
| QR | 31 (41.3%) | 21 (28%) | 2.258 | 1.118-4.558 | 0.023* |
| RR | 10 (13.3%) | 2 (2.67%) | 7.647 | 1.577-37.071 | 0.012* |
| Allele Frequency |
| Q | 0.66 | 0.833 | - | - | - |
| R | 0.34 | 0.167 | - | - | - |
Tables show the prevalence of various genotypes in mothers (a) and in neonates (b). Also, shown is the odds ratio (using Chi square test) of incidence of IFGR with the specific genotype. Allele frequency for each allele is shown for both mothers and neonates of both groups separately. *p<0.05 was considered to be significant.
Gene Expression
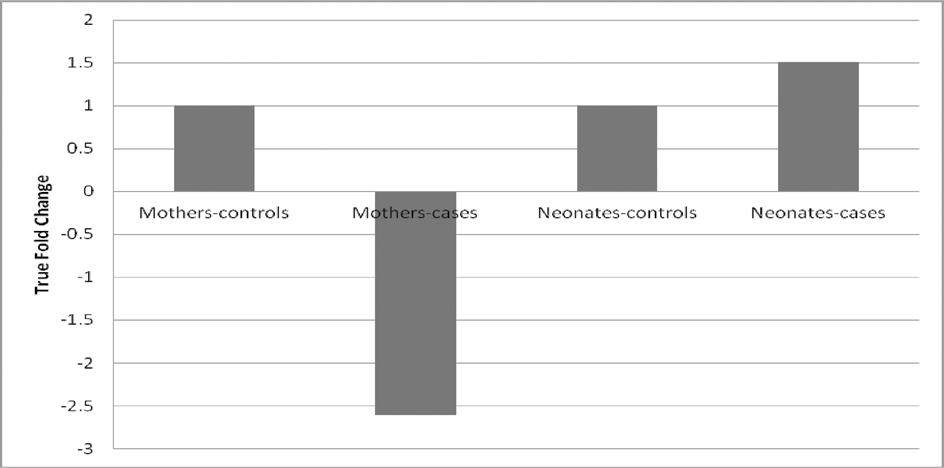
Reverse Transcription- quantitative (real time) polymerase chain reaction, which was used for estimating the expression levels of PON1 mRNA in the blood samples of the study subjects, showed differing results in mothers and neonates. Change in ΔCt value (ΔΔCt) was negative in mothers (-1.38), which indicated a decrease in expression of mRNA in mothers giving birth to IFGR neonates, while a positive ΔΔCt in foetus (0.59) is indicative of increase in expression of mRNA in IFGR neonates. To find out the degree of change in expression, True FC was calculated.
As can be seen from [Table/Fig-5], there was decrease in mRNA levels of the PON1 gene in cases of mothers as compared to controls, and the decrease was 2.6 times that of the control group. However, the foetus group showed increased PON1 mRNA expression in the cases group when compared with the control group, and the increase was 1.5 times more.
True Fold Change in mothers and neonates for PON1 mRNA expression. Shown here is the difference in expression of PON1 in mothers (cases) and neonates (cases) as compared with controls. A decrease in expression is seen in mothers, while expression in neonates was found to be increased.
Biochemical Activity
PON1 levels [Table/Fig-6] in mothers giving birth to IFGR babies was about 30% less than the values found in the control group of mothers (118 vs 147.77 mol min-1 mL-1 of serum), and this difference was found to be statistically significant (p<0.05). However, similar results were not found in the foetal plasma samples. The PON1 enzymatic activity that was the calculated in the IFGR babies was found to be 127% higher than control group (123.49 vs 54.74 mol min-1 mL-1 of serum), and this difference was also found to be statistically significant (p<0.001).
Shown here are the levels of enzymatic PON1 activity in the various groups. PON1 enzymatic activity was found to be decreased in mothers of IFGR neonates, while the values were found to be increased in the IFGR neonates. The differences in both mothers and neonates were found to be statistically significant (p<0.01 for both groups).
| Groups | PON serum enzyme activity | S.D. |
|---|
| Mothers- controls | 147.77 | 77.83 |
| Mothers- cases | 118 | 45.54 |
| Neonates- controls | 54.74 | 28.92 |
| Neonates- cases | 123.49 | 68.26 |
Correlation Analyses
Correlation analysis was carried out between foetal weight and serum PON1 levels to determine the trend of change of foetal weight with this parameter and whether, the change was statistically significant or not. There was no statistically significant correlation of maternal PON1 levels with foetal weight (r=0.068; p=0.4) but significant negative correlation of foetal levels of PON1 with foetal weight (r=-0.195; p=0.016) was observed.
On statistical analysis, the difference in activity of PON1 was found to correlate significantly with the genotype (R allele resulting in lower activity) in mothers giving birth to IFGR neonates (p-value=0.042). Although, the pattern of alteration of activity in relation to genotypes was similar in the other three groups, the correlation was statistically not significant (p-value: mothers (control)=0.1; IFGR neonates=0.07; neonates (control)=0.387).
Discussion
The aim of this study was to examine the relationship of IFGR with PON1 gene polymorphism and expression in both neonates and mothers. Our results showed that PON1 Q192R polymorphism is associated with IFGR. We also found a significant impact of PON1 expression in mothers on foetal weight, an important risk marker for IFGR. Considering the distinct ethnic feature of the Indian population, the high prevalence of IFGR in them and the absence of any report on PON1 gene polymorphism in Indian IFGR patients, the findings of this report are of considerable importance.
Genotype Prevalence
In this study, the prevalence of the PON1 Q192R genotype was found to be as 62% (QQ), 34.7% (QR) and 3.3% (RR) in the mother and foetus controls. In a separate study, different distributions of genotypes were found in different populations. The distribution of genotypes in Finnish and American Caucasian population was found to be 50-52% (QQ), 41-44% (QR) and 4-9% (RR). Thus, the prevalence in our study was similar to American Caucasian and European population. However, the frequencies in Indian population in same study were reported as 47% (QQ), 40% (QR) and 13% (RR), which were quite different from the other Asian populations. The reported frequencies of PON1 Q192R genotypes in Japanese, Chinese, Malay and American Blacks population were 15-24% (QQ), 35-50% (QR) and 33-42% (RR). The results of genotype frequency in our study are quite different from the previous reports [23]. This difference is probably because of difference in case selection; while the previous study included subjects from both North and South India, subjects in our study were drawn only from North Indian population, in and around Delhi. The difference is therefore expected, since ethnic variation between North and South Indian population has already been established [24].
In mothers of IFGR neonates, the prevalence of QR genotype was found to be 76% and that of RR genotype was 8%. Mothers having R allele were found to have higher odds ratio of development of IFGR [Table/Fig-4a]. High prevalence of QR and RR genotype were also associated with low level of PON1 expression and activity. This low level of PON1 activity arising due to genetic polymorphism may be indirectly responsible for development of IFGR. In the neonates also, the presence of R allele was found to be associated more commonly with development of IFGR. No significant report is available regarding PON1 polymorphism in IFGR materno foetal dyads to compare the result.
Gene Expression
The variation in PON1 expression in the study subjects was estimated by measuring the mRNA levels. The levels between cases and controls were compared for materno-foetal dyads. A decrease in expression levels was found in mothers giving birth to IFGR babies as compared to controls. This lower expression corroborates with the low plasma PON1 activity in mothers of IFGR babies. This is consistent with the accepted knowledge that inflammatory state gives rise to increased Oxidative Stress (OS) and inflammatory state causes decrease in levels of PON1 [25,26] and hints at the possibility that an underlying inflammatory state may be playing a role in IFGR along with the decreased antioxidant capacity of the subjects.
Biochemical Activity
Decreased PON1 levels in the maternal serum can also lead to placental insufficiency. Chen D et al., have postulated that the milieu of increased OS and decreased PON1 levels may lead to formation of microatheromas in the placental microvasculature, leading to blockage of these vessels due to increased oxidation of LDL [11]. This, in turn, will give rise to placental insufficiency leading to IFGR. However, in IFGR babies, an increase in PON1 expression levels was found when compared with controls. This finding can be explained by the influence of adipose content of the body on PON1 levels. Adipose stores in the body are inversely related to the serum PON1 levels; lower the adipose stores of the body, more will be the PON1 levels in the serum. Thus, in FGR neonates, where the body stores of adipose tissue is lower when compared with the controls, the higher PON1 levels in the serum are expected.
While the PON1 activity in IFGR babies was found to be significantly increased, it was found to be significantly decreased in their mothers, as shown in [Table/Fig-6]. This finding is in agreement with the results obtained by the expression study (RT-PCR) and helps to re-establish the presence of higher levels of PON1 enzyme in the IFGR neonates, which may be caused by decreased adipose stores of the body, as discussed previously [27].
Paraoxonase enzyme has been shown to be an important antioxidant in the serum and most of the esterase activity of HDL is ascribed to the PON1 enzyme attached to HDL [28]. It has been definitively shown that the reduction of oxidised LDL is a result of the arylesterase activity of PON1. Also known is the fact that oxidised LDL is an important precursor to plaque formation, which eventually leads to clotting and clogging of vessels. It has been postulated that decreased PON1 activity in mothers may promote blockage of placental vessels, thus, inducing ischemia reperfusion type of injury in addition to the decreased antioxidant capacity of the body, thus, leading to increased OS and amplification of the effects of its decreased activity [29].
In this study, the activity of PON1 in plasma was significantly decreased in mothers giving birth to IFGR babies. PON1 activity however, was not found to correlate significantly with foetal weight. Similarly, a study involving Korean women with preterm delivery also showed no significant difference in serum PON1 levels [30]. The levels of PON1 activity in the IFGR babies themselves were found to be conversely elevated. PON1 enzymatic activity and foetal weight was found to be negatively correlated (r=-0.195, p=0.016). Foetal PON1 enzymatic activity has shown significant increase with increase in OS and decrease in foetal weight.
Limitation
The extrapolation of any conclusions from the present study may be insufficient on account of small sample size.
Conclusion
In conclusion, the results of the present study demonstrate that pregnant females with mutant variants of Q192R polymorphism associated with low PON1 expression and activity in mothers are at a risk of giving birth to IFGR neonates. Further large, multiethnic studies should be performed to confirm the role of PON1 gene polymorphism on susceptibility to IFGR neonates in pregnant females.