Students living in hostels, military recruits etc., are considered to be at a higher risk of acquiring meningococcal disease as they live in close proximity and as such are strongly recommended vaccination in the developed countries like the United States and many European countries [1,2].
The epidemiology of meningococcal disease is not well described in many parts of the developing world, particularly Asia including India [4,5]. Sinclair et al., reviewed published literature from India and concluded that there is a low background incidence of meningococcal disease with occasional large epidemics [6]. In the absence of adequate surveillance for meningococcal disease, many additional smaller outbreaks have gone unnoticed and the size of the reported ones underestimated. Because carriage of meningococci is much more frequent than invasive meningococcal disease [1,2], the epidemiology of meningococcal infection cannot be understood without studying the meningococcal carriage. Studies on carriage can identify demographic and socio-behavioural risk factors for meningococcal infection.
Against this backdrop a study to assess the nasopharyngeal carriage rate among university students residing in hostels would shed some light on whether Indian university students residing in hostels are indeed at risk of suffering from invasive meningococcal disease like their counterparts in the US and Europe [4]. Moreover, this study would add to the scarce literature on nasopharyngeal carriage in any section of the Indian population. The present study was thus designed to ascertain the nasal carriage of meningococci among college freshmen in a temperate north Indian setting.
Materials and Methods
The cross-sectional study was conducted in National Institute of Technology, Srinagar in the north Indian state of Jammu and Kashmir from August to September 2014. Two hundred seventy-four engineering college freshmen who were first year students residing in a single hostel were recruited for the study. Informed consent was obtained from all the college freshmen who joined the college hostel for the first time and the participants (aged >18 years) were asked to provide some basic information regarding demographic profile, history of any high risk behaviours, recent/past illness, intake of steroids or any other immunosuppressant, vaccinations or recent antibiotic use. Students who were vaccinated for meningococcal disease, those with a history of antibiotic intake, use of immune suppressants or steroid drugs in past 2 weeks or any immunological or haematological disorder were excluded from the study.
A charcoal impregnated posterior pharyngeal swab was taken from each student fulfilling the inclusion criteria and transported to the laboratory within 2-3 hours and plated on Thayer Martin medium. The plates were incubated at 370C with 5-10% CO2 for 24 hours at the end of which they were examined for any growth. In case of no growth, the plates were incubated for another 24 hours and examined subsequently. Oxidase positive Gram negative diplococci suggestive of being Neisseria spp. were sub-cultured and tested for molecular identification and sequence analysis.
Identification by sequencing of 16S rDNA: Genomic DNA from the bacteria was isolated according to the protocol of Kumar et al., [7]. Amplification of 16S rRNA gene was performed on master thermal cycler (UK), with universal primer set pA (forward primer) (5’-AGA GTT TGA TCC TGG CTC AG- 3’) and pH (Reverse primer) (5’-AAG GAG GTG ATC CAG CCG CA- 3’) [8,9] in 25 μl of reaction containing 1X Taq buffer, 100 μMol l-1dNTPs mix, 3 mMol l-1 MgCl2, 10 μg BSA(bovine serum albumin), 10 pMol each primer, 0.5 U of Taq DNA polymerase and 100 ng template DNA. The thermo cycling conditions consisted of an initial denaturation at 94°C for 2 minutes, 35 amplification cycles at 94°C for 1 minute 10 seconds, 48°C for 30 seconds, 72°C for 2 minutes 10 seconds and a final polymerization step at 72°C for 6 minutes 10 seconds. The final PCR product was resolved in 1% agarose gel, excised and purified with Sigma elution kit. The cycle sequencing reaction was performed with 20-30 ng of purified amplicon using the ABI PRISM BigDye Terminators v1.1 cycle sequencing kit according to the manufacturer’s instruction (Applied Biosystems Foster city, CA, USA). The purified product was sequenced bi-directionally on AB3100 sequencer (Applied Biosystems) to obtain complete coverage of the gene. The sequences were edited, contig assembled in CLC Bio Sequence viewer and compared with GenBank sequences by blast analysis. Nucleotide sequence similarities were determined using the NCBI or EMBL databases and sequence identity vis-à-vis the bacterial identity was established by closest match [10].
Sequence and molecular Phylogenetic Analysis: Blast analysis was performed first against the retrieved sequences from the whole NCBI nr/nt database and subsequently against each other to analyse the differences in the dataset. ClustalW2 was used for multi sequence alignment (MSA) to see the conserved regions among the sequences, MEGA 6 package were used for molecular phylogenetic analysis. Fitch cost estimation was done and on the basis of that agglomerative clustering was done using the “bottom up” approach. Implementations and performance of hierarchical clustering was performed to infer the complex phylogenetic representations in the context of N. meningitidis observed data. On the basis of blast identification the sequenced samples were assigned SKIMS ID, Blast Identification, Sequence ID, Blast score and E-Value. Operational Taxonomic Unit numbers (OTU) were also assigned for molecular phylogenetic analysis. DNA sequences were deposited in the GenBank database with the accession numbers as shown in [Table/Fig-1]. To identify the particular serogroup of N.meningitidis out of dataset1, by using Blast N, different N.meningitidis sero groups were retrieved from NCBI database and from 16s database RDPII to develop dataset2 that included the pathogenic Neisseria sequences of dataset1 and retrieved N.meningitidis serogroups [Table/Fig-2]. Substitution model was estimated to determine the best mutation substitution model fit for the dataset. Maximum Likelihood (ML) estimation of substitution matrix and gamma parameter was performed. Rates of different transitional substitutions and those of transversional substitutions are shown in [Table/Fig-3]. Tree topology was automatically computed for estimating ML values. The maximum log-likelihood for this computation was -704.887. The analysis involved 26 nucleotide sequences (dataset2). Areas containing missing data and gaps were eliminated. There were a total of 69 positions in the final dataset.
Dataset with blast identification and GenBank accession numbers.
| NO | Identification (BLAST) | SKIMS Sequence ID | GenBank Accession Numbers | BasePair | Percentage Similarity | Score | E-Value | OTU1 |
|---|
| 1 | Neisseria Sp. | Skimsmen0001 | KR870878 | 281bp | 94% | 394 | 4E-106 | 1 |
| 2 | Neisseria perflava | Skimsmen0002 | KR870879 | 1489bp | 99% | 2538 | 0.0 | 2 |
| 3 | Neisseria meningitdis | Skimsmen0003 | KR870880 | 1475bp | 99% | 2597 | 0.0 | 3 |
| 4 | Neisseria pharynges | Skimsmen0004 | KR870881 | 1476bp | 97% | 2422 | 0.0 | 4 |
| 5 | Neisseria flavescens | Skimsmen0005 | KR870882 | 1481bp | 99% | 2623 | 0.0 | 5 |
| 6 | Neisseria perflava | Skimsmen0006 | KR870883 | 1473bp | 99% | 2634 | 0.0 | 6 |
| 7 | Neisseria meningitidis | Skimsmen0007 | KR870884 | 1473bp | 96% | 2603 | 0.0 | 7 |
| 8 | Neisseria perflava | Skimsmen0008 | KR870885 | 1478bp | 99% | 2595 | 0.0 | 8 |
| 9 | Neisseria meningitidis | Skimsmen0009 | KR870886 | 1475bp | 99% | 2603 | 0.0 | 9 |
| 10 | Neisseria meningitidis | Skimsmen0010 | KR870887 | 1475bp | 99% | 2603 | 0.0 | 10 |
List of retrieved Neisseria meningitidis serogroups from NCBI & RDPII databases (data set2).
| S no. | GenBankAccession no. | Species | Serogroups | OTU1 no |
|---|
| 1 | gi|2425160 | Neisseria Meningitidis | Serogroup W135 | 11 |
| 2 | gi|578002988 | Neisseria Meningitidis | Serogroup Y | 12 |
| 3 | gi|578002986 | Neisseria Meningitidis | Serogroup C | 13 |
| 4 | gi|352289265 | Neisseria Meningitidis | Serogroup D | 14 |
| 5 | gi|78522969 | Neisseria Meningitidis | Serogroup B | 15 |
| 6 | gi|78522967 | Neisseria Meningitidis | Serogroup B | 16 |
| 7 | gi|78522931 | Neisseria Meningitidis | Serogroup B | 17 |
| 8 | gi|78522929 | Neisseria Meningitidis | Serogroup B | 18 |
| 9 | gi|78522951 | Neisseria Meningitidis | Serogroup B | 19 |
| 10 | gi|78522965 | Neisseria Meningitidis | Serogroup B | 20 |
| 11 | gi|78522933 | Neisseria Meningitidis | Serogroup B | 21 |
| 12 | gi|78522959 | Neisseria Meningitidis | Serogroup B | 22 |
| 13 | gi|78522953 | Neisseria Meningitidis | Serogroup B | 23 |
| 14 | gi|78522961 | Neisseria Meningitidis | Serogroup B | 24 |
| 15 | gi|78522957 | Neisseria Meningitidis | Serogroup B | 25 |
| 16 | gi|30407145 | Neisseria Meningitidis | Serogroup A | 26 |
1OTU No: Operational Taxonomic Unit Number
Shows transitional & transverstional substitution rates.
| A | T/U | C | G |
|---|
| A | --- | 6.36 | 6.36 | 12.28 |
| T/U | 6.36 | --- | 12.28 | 6.36 |
| C | 6.36 | 12.28 | --- | 6.36 |
| G | 12.28 | 6.36 | 6.36 | --- |
Evolutionary analyses were conducted in MEGA6. On the basis of estimated substitution parameters, different phylogenetic trees were constructed for different data sets to ascertain the similarity of the N.meningitidis in the current study vis a vis retrieved sequences of serogroup B. After performing sequence analysis, MSA different pathogenic N. meningitidis serogroups A, B, C, D, Y & W125 were identified and then retrieved from NCBI. These sequences were subjected to phylogenetic analysis.
The study protocol was approved by the Institute Ethics Committee of Sher-i-Kashmir Institute of Medical Sciences. Informed consent was obtained from all participants. Descriptive statistics employing SPSS (Statistical Package for Social Sciences) Ver 17.0 (IBM, USA) was employed for the data analysis.
Results
Of the 274 samples, 10 (3.6%) grew Gram-negative diplococci suggestive of Neisseria spp. Molecular identification revealed the various isolates to be homologous to N.meningitidis (n=4), N. perflava(n=3), N.flavescens, N. pharynges and N.spp(1 each) [Table/Fig-1]. The length of the sequences ranged from 281to 1489bp.
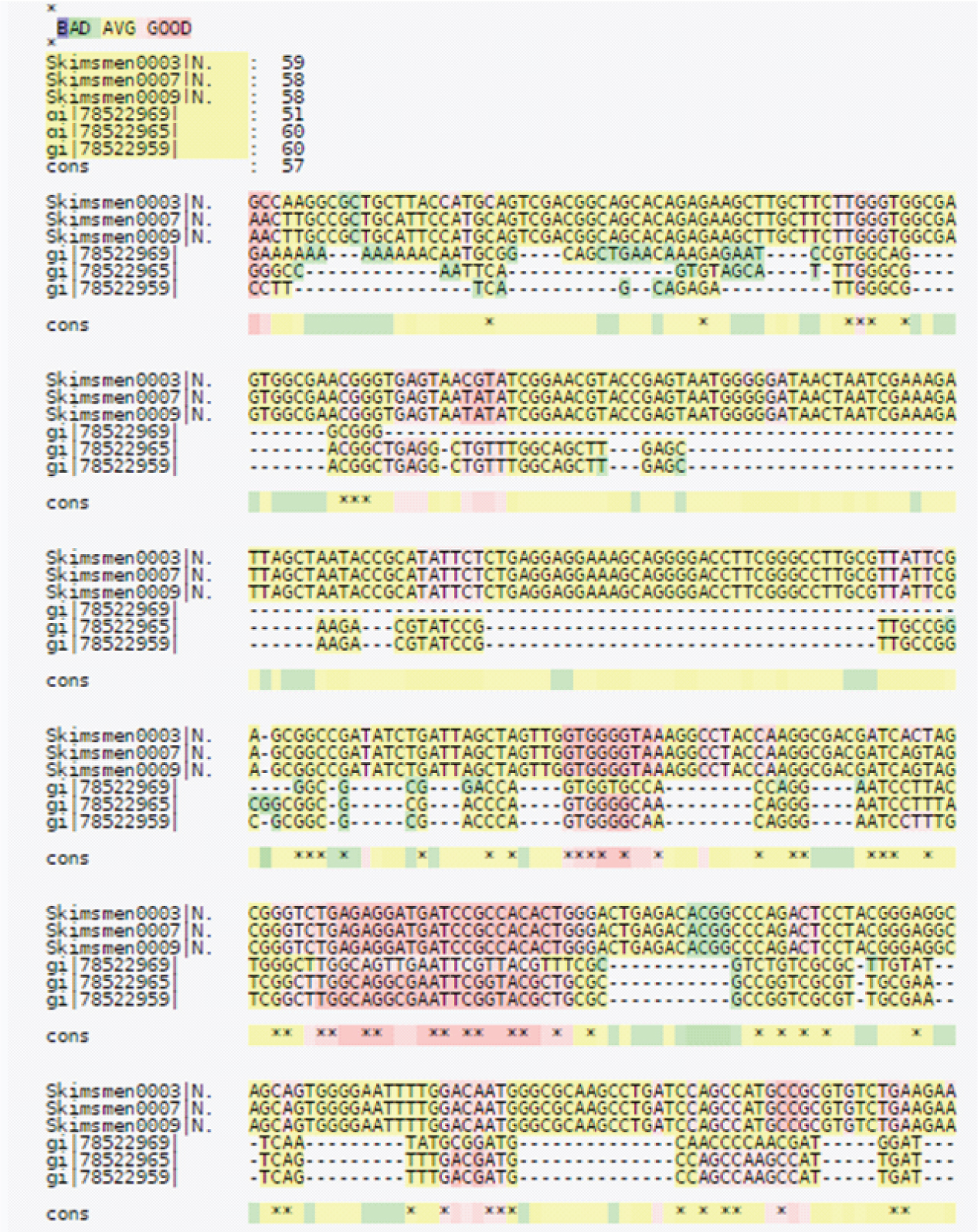
In the dataset, the insertion/deletion or a single base differentiating the dataset into pathogenic or non-pathogenic species showed that 4 sequences (ID’s SKIMSMEN0003, SKIMSMEN0007, SKIMSMEN0009 and SKIMSMEN0010) showed 99% identity with pathogenic N. meningitidis on different E-value parameters. The sequences were compared to one another by MSA. On the basis of sequence similarity, total score and E-value the 16S rRNA sequences belonged to different Neiserria species shown in [Table/Fig-1]. Multiple sequence Alignment showed conserved regions among the sequences. Sequence ID’s SKIMSMEN0003, SKIMSMEN0007, SKIMSMEN0009 and SKIMSMEN0010 were found to be highly conserved among the residues. From sequence alignment Sequence ID’s gi|78522959, gi|78522969 & gi|78522965 showed some mutations and variations in the residues, as sequence similarity showed a matching similarity of 51%, 58%, 58%, 51%, 59%, 60%, 60% & 57% respectively [Table/Fig-4].
Multiple sequence alignment of SKIMSMEN0003, SKIMSMEN0007, SKIMSMEN0009 and SKIMSMEN0010 gi|78522969, gi|78522965, gi|78522959&gi|78522953.
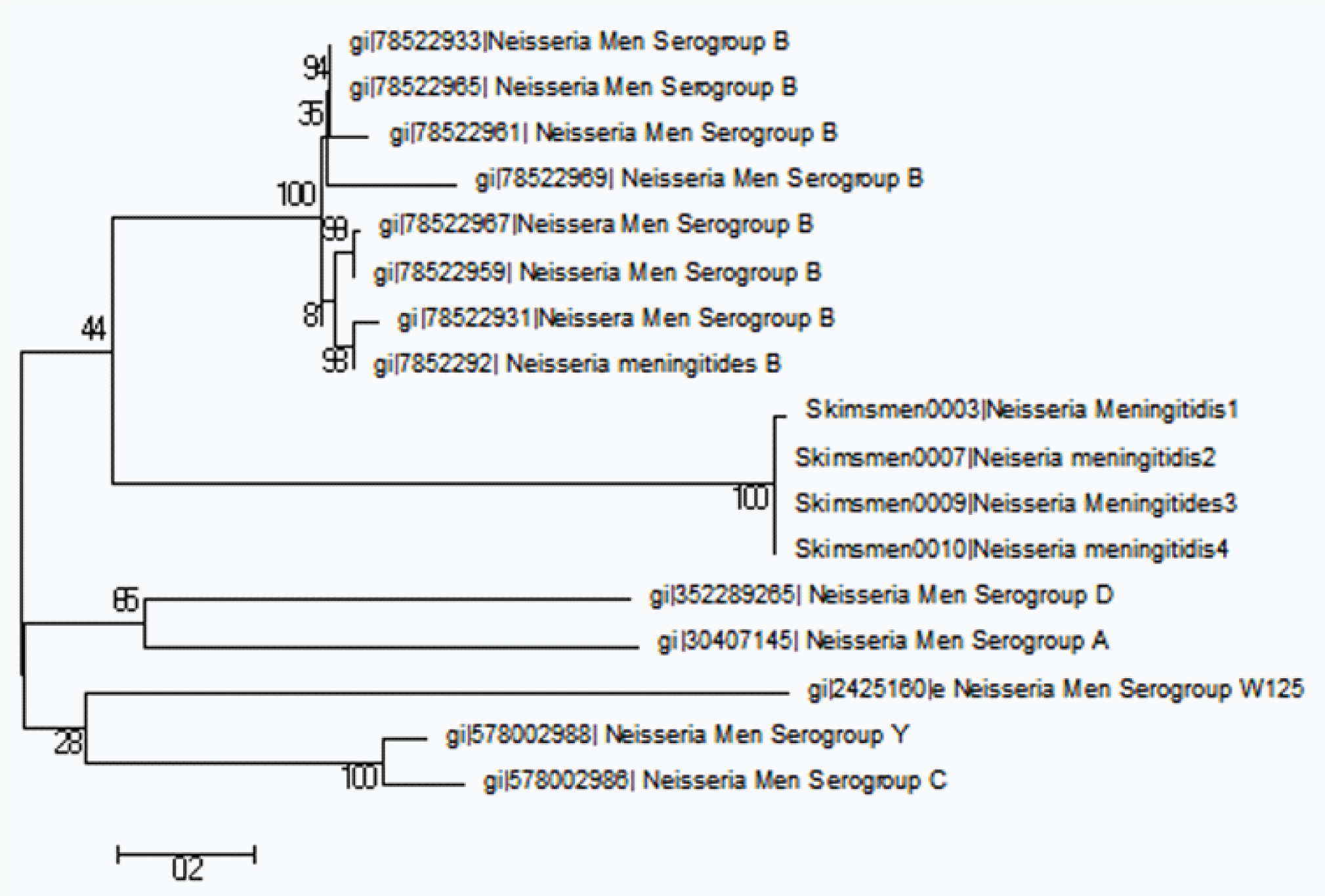
After performing sequence analysis and MSA of different pathogenic N. meningitidis phylogenetic analysis was performed with various retrieved serogroups. On phylogentic analysis N. meningitidis serogroup with ID gi|78522959 showed close relationship with sequences of ID’s SKIMSMEN0003, SKIMSMEN0007, and SKIMSMEN0009& SKIMSMEN0010. Serogroup D & A were distinctly related whereas Serogroup Y & C were found to be distantly related as shown in [Table/Fig-5]. Model substitution identification analysis revealed that Kumara-2-parametre model was the best fit for the dataset. Clustering analysis was done to infer maximum phylogenetic representations on the basis of Fitch cost. It was seen that OTU 15 (Genbank Id gi|78522969): N.meningitidis serogroup ‘B’ aligned closely with N.meningitidis sequences in dataset1 followed by other retrieved serogroup B sequences [Table/Fig-6]. OTU 26 (Genbank Id gi|30407145: Serogroup ‘A’) and OTU 14 (Genbank Id gi|352289265: Serogroup ‘D’) were observed to be distinct compared to OTU 11 (gi|2425160: Serogroup ‘W125’), OTU13 (Genbank Id gi|578002986, Serogroup ‘C’) and OTU 12 (Genbank Id gi|578002988, Serogroup ‘Y’) [Table/Fig-6].
Shows the evolutionary relationship of different serogroups of N.meningitidis by using Mega 6.
Inference of multiple phylogenetic representations by clustering using fitch metric and the average Linkage Criterion.
1H: Hypothetical Taxonomic Unit
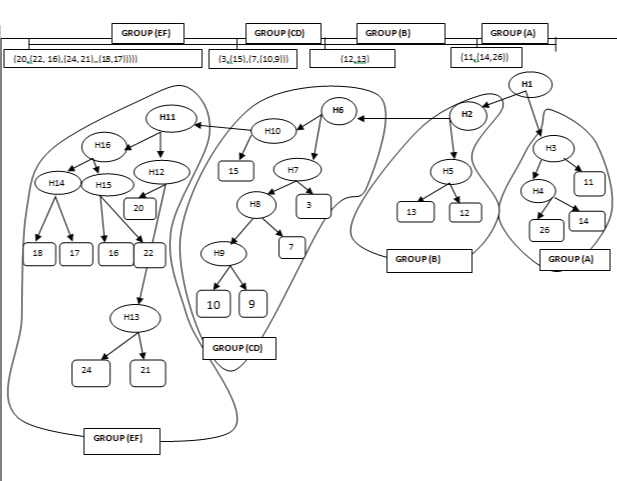
Phylogenetic trees revealed the similarity of the N.meningitidis in the current study with retrieved sequences of serogroup B [Table/Fig-6]. Agglomerative clustering using Fitch metric and the average linkage criterion produced a constant result which appeared to have an optimal cost by Fitch and by the combined metric. Four groups illustrated by their OTU numbers were observed that were identical in all most all phylogenetic representation and included Group A: (11,(14,26))); Group B: (12,13); Group CD: (3,(15,(7,(10, 9)))) and Group EF: (20,(22,16),(24,21),(18,17)))). We got multiple phylogenetic representations from different datasets on different parameters. To infer such big data clustering analysis was done to merge all relevant OTUs on the basis of Fitch cost. The identified pathogenic taxonomic units viz., OTU 3, OTU 7, OTU 9 and OTU 10 shared the common Clade H6 (HTU1) that clubbed with the OTU 15 which is serogroup ‘B’ and formed “Group CD” as shown in [Table/Fig-6]. Hypothetical Taxonomic units (HTU) 3 and HTU 4 got clubbed in a single “Group A” including OTUs 11, 14 and 26. OTU 12 and OTU 13 were found to get clubbed with “Group B”. OTUs 20, 22, 16, 17, 18, 21 and 24 shared the common HTU: H11, all OTUs got clubbed into “Group EF” as shown in [Table/Fig-6].
Discussion
Regardless of age, N.meningitidis is responsible for 1.9% of cases of meningitis in India [6] affecting adults as well as children [11-13]. While mostly children are affected with invasive disease, outbreaks witness affliction of adults as well [6]. Our data show that N.meningitidis Serotype Bis carried in the nasopharyngeal swabs of 1.5% college freshmen. This is the first study from our part to estimate the prevalence of the organism in college students in India and should serve as an impetus for further such studies on similar participants who live is close circumstances.
Our data is in conformity to earlier reports of a lower meningococcal carriage in Indian populations than that reported in other settings, even during epidemics, and without obvious age or sex predilections [14-20] [Table/Fig-7]. During the 1985 epidemic in New Delhi, the prevalence of carriage in healthy subjects aged 6–20 years was below 2% [13]. These findings were consistent with the rather low level of carriage seen in in-coming prisoners and army recruits in previous studies [14,21]. These populations serve as proxy indicators of adult population and point to a general lower nasopharyngeal carriage of meningococci in Indian adults [14,16,21]. Higher carriage rates have been found in close family contacts of cases [15,16,18]. The effect of staying in close settings on the prevalence of nasal carriage needs to be studied which might dictate the usefulness of targeted chemoprophylaxis. The study employed 16sRNA gene sequence analysis for molecular and serogroup identification of N.meningitidis. Determination of differences in the sequence of the 16S rRNA gene is well established as a standard method for the identification and phylogenetic classification of prokaryotic species, genera, and families [22]. This is rapidly becoming a common technique for the identification of unknown bacterial isolates and we employed this molecular method of bacterial identification for the first time in our valley.
Summary of reported Indian series of nasopharyngeal carriage of meningococci. (modified from Sincliar et al., [6]).
| Study | Site/year | Study population | Carriers | Comment |
|---|
| Ardeshir [14] | Nasirabad 1932/1933 | 375 new recruits 400 staff members Recruits on leaving | 7 (1.8%) 176 (44%) | Army training centre |
| Bhalla et al, [15] | Hyderabad 1968 | 55 close contacts | 3 (5%) | A military training centre (serogrouping not available) |
| Ichhpujani et al., [16] | New Delhi 1985 | 242 family contacts 213 neighborhood contacts 160 school contacts | 42 (17.4%)16 (7.5%)1 (0.6%) | All were serogroup A. |
| Dubey et al., [17] | Chandigarh 1986 | 32 close contacts | 11 (34.3%) | Carrier rate during non-epidemic period in Chandigarh 3%. Same serotypes as seen in cases (A or C) |
| Paul et al., [18] | New Delhi 1985 | 53 household/family contacts 219 neighborhood/ school contacts | 3 (5.7%)2 (0.9%) | Serogroup Aserogroup A (n=1), C (n=1) |
| Ichhpujani et al., [19] | New Delhi 1986/1987 | 6513 school children (aged 6–20 years) | 107 (1.64%) | No differences in sex or age groups. |
| Nagraj et al., [20] | Baddi village 1997 | 25 close contacts of suspected cases | 0 (0%) | Nearly all had received chemoprophylaxis. |
| Present study | Srinagar(2014) | 274 college freshmen | 4 (1.5%) | Serogroup B |
Our data is in agreement with earlier studies that have reported a lower meningococcal carriage in Indian populations than those reported in other settings, even during epidemics [Table/Fig-7]. Most of the cases are caused by a few clonal complexes of related sequence types commonly known as hyper virulent lineages as revealed by multi-locus sequence typing [23] and isolates belonging to these hypervirulent lineages are under-representation in healthy carriers [24,25]. In turn, among the healthy population only 10% are colonized with N. meningitidis and most of these isolates belong to a diverse variety of 2,000 sequence types that are rarely associated with disease [26]. In our study also we found that 60% of the isolates belonged to non-pathogenic sequence types rather than the pathogenic N.meningitidis [Table/Fig-1].
We found that the menincococcal sequences of our strains aligned with serogroup B. Meningococci can be divided into 13 serogroups based on the chemical composition of the capsular polysaccharide, invasive disease being largely limited to serogroups A, B, C, W, X, and Y. Our data are at variance with the earlier Indian reports, predominantly related to outbreaks, where the serogroup A has been the commonest reported serogroup [6,12,13,27,28]. There have been rare reports of disease attributable to serogroup C [6] and one fatal case of infection with serogroup B has been reported [29].
Indians are not routinely advised meningococcal vaccination, it being reserved for pilgrims prior to Hajj [30] and army cadets on overseas assignments [31]. While the low level carriage in our data would argue against routine vaccination, similar studies across other high risk groups and ages need to be performed before conclusive recommendations for vaccinations can be made. Further geographical differences in the carriage could occur in a large country like India where more outbreaks have been reported from temperate northern areas of the country as compared to the more tropical Southern areas [6]. Assessment of the community burden would dictate concrete vaccination recommendations as also determine whether the commonly available meningococcal vaccine needs to incorporate a particular serogroup for effective protection.
Limitation
Our study is limited by the fact that it was carried out in only one north Indian setting and the results are not generalizable to the whole country. Further the effect of living in close proximity was not studied which would involve a repeat survey of the studied students after 6 weeks but was not possible as the institutions were closed following untimely flash floods.
Conclusion
We conclude that nasal carriage of N. meningitidis serogroup B was seen in 1.5% of fresh college recruits in our state. Whereas earlier reports from different parts of the country carried out during epidemics have implicated serogroup A and C as the predominant serogroups causing disease, our study calls for similar studies from other areas of the country in order to assess the carriage in those areas and the relevance of the vaccine as regards the serogroup.
Data Guarantor
All the authors stand as guarantors of the data reported in the study.
Financial Disclosures
The study was supported in part by Sanofi India Pvt Ltd as an investigator initiated study. The funding source had no role in the design or conduct of the study or in the analysis and interpretation of the resulting data.
1OTU No: Operational Taxonomic Unit Number