Urine is the most common type of specimen received by clinical microbiology laboratories [1]. Useful diagnostic tools for UTI include the urine dipstick test and microscopic examination, both of which provide point-of-care information, and urine culture, which is considered as the gold standard for the diagnosis of UTI [2]. Proteinuria is defined as excretion of more than 300 mg albumin per day [2]. UTI is often associated with proteinuria [3,4], however, relationship between proteinuria and UTI remains incompletely understood [4]. As many as 60%-80% of all urine specimens received for culture contain no aetiological agent or contain only contaminants [1]. While urine culture is cumbersome, time consuming and expensive; screening procedures on the other hand are simple to perform, rapid and inexpensive [5].
We undertook this study with the objectives: 1) To find out the association between proteinuria and UTI and; 2) Whether proteinuria could be a guide to empirical therapy for UTI among proteinuria positive and negative cases.
Materials and Methods
A prospective study was conducted at the Microbiology Department of Dr. Baba Saheb Ambedkar Hospital, New Delhi, India, over a period of three months (April 2015 to June 2015).
Inclusion and Exclusion criteria: All consecutive urine specimens (both catheterized and noncatheterized urine samples) for which both routine microscopy and culture was requested, received in the Microbiology Department between April 2015 to June 2015 were included in the study. Any repeat specimen from the same patient received during the three months period was excluded from the analysis.
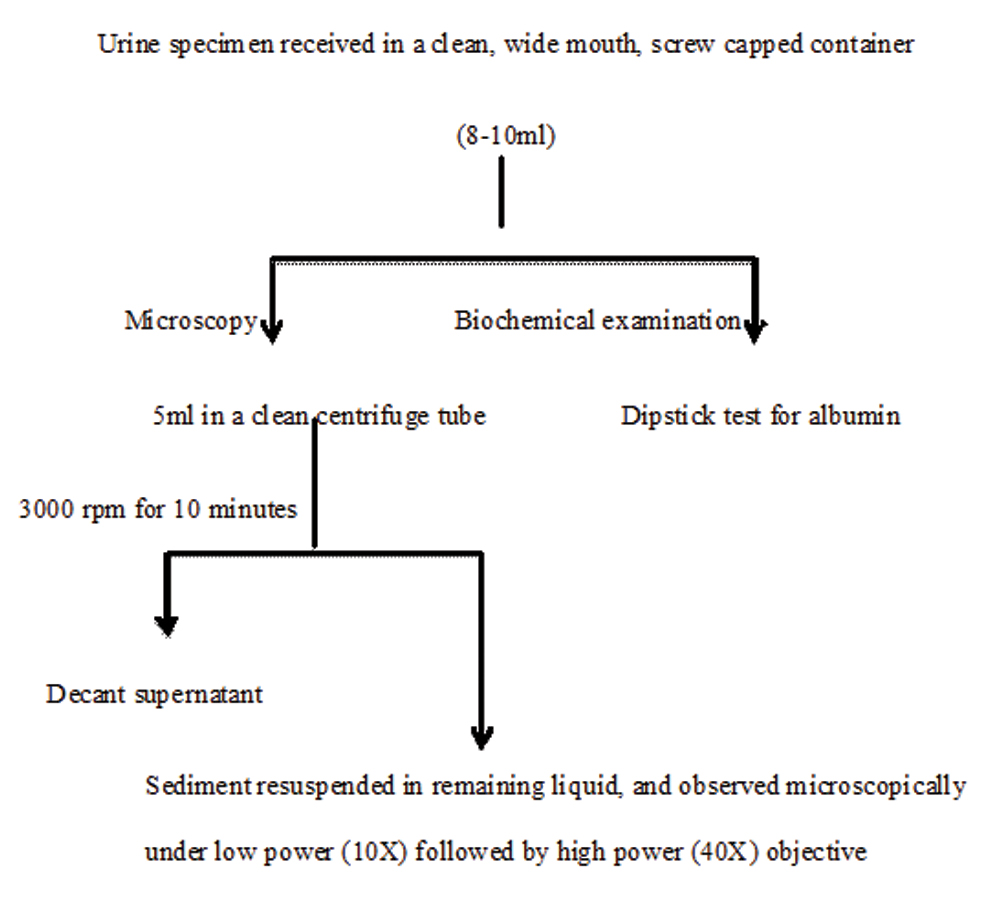
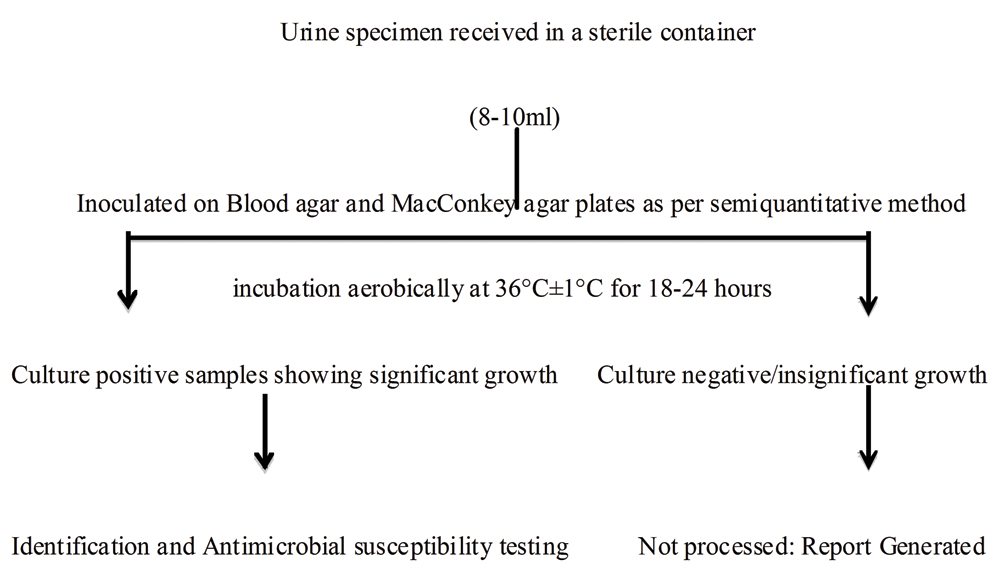
Specimen processing: Urine specimens received in the microbiology laboratory were analysed microscopically and by dipstick test [Table/Fig-1]. The urine sediments were analysed microscopically under low power (10X) followed by high power objective (40X) for presence of polymorphonuclear leucocytes, red blood cells, bacteria, fungi, cast and crystals. Semiquantitative estimation of urinary albumin was done using Accu-Stix (Accurex Biomedical Pvt. Ltd., Mumbai). It is a reagent-strip test, based on the principle ‘protein-error of-indicator’ for estimation of urinary protein. Urine routine and microscopy findings were compared to urine culture results of a few representative samples, for which a separate urine specimen along with requisition of culture and antibiotic susceptibility testing was received in the department. Specimens for urine culture were inoculated on blood agar and MacConkey agar plates as per semiquantitative method, using standard loop of 4 mm diameter [1]. The plates were incubated aerobically at 36°C±1°C for 18-24 hours. The culture result was interpreted as per Kass criteria of significant bacteriuria [1]. Isolates were examined for colony characteristics, Gram staining, motility and biochemical tests [Table/Fig-2].
Flowchart depicting processing of urine specimen for routine and microscopic examination.
Flowchart depicting processing of urine specimen for culture and antimicrobial susceptibility testing.
Antimicrobial susceptibility testing: Culture positive isolates showing significant growth were processed further. Antimicrobial susceptibility was performed by Kirby-Bauer Disc diffusion method and the results were interpreted as per CLSI guidelines [6]. The antibiotic discs tested were chloramphenicol (30 μg), gentamicin (10 μg), ceftriaxone (30 μg), cefuroxime (30 μg), piperacillin-tazobactam (100/1 μg), ampicillin (10 μg), amoxicillin-clavulanic acid (20/10 μg), cotrimoxazole (23.75/1.25 μg), ciprofloxacin (5 μg), linezolid (30 μg), teicoplanin (30 μg), imipenem (10 μg), vancomycin (30 μg), clindamycin (2 μg), levofloxacin (5 μg), amikacin (30 μg), nalidixic acid (30 μg), cefotaxime (30 μg). Escherichia coli (ATCC® 25922TM), Klebsiella pneumoniae (ATCC® 700603TM), Pseudomonas aeruginosa (ATCC® 27853TM) and Staphylococcus aureus (ATCC® 25923TM) were used for quality control [6].
Statistical Analysis
Statistical analysis was done by Fisher’s-exact test and p-value was calculated using two by two table. A p-value ≤0.05 was considered as significant.
Results
A total of 369 (study group) urine specimens were received for routine and microscopic examination and culture during the study period. Out of these 73 came out to be proteinuria positive while 296 were proteinuria negative. Among the proteinuria positive cases, 52.7% were from patients visiting outpatient department while 47.3% were from patients admitted in various wards including surgical wards (10.7%), medical wards (34.2%), obstetrics and gynaecology ward (1.4%), paediatric wards (48.5%) and ICU (5.2%). Out of 73 proteinuria positive cases, 32 were found to be culture positive with significant growth while 41 were culture negative. Patient characteristics and urinary parameters were analysed in proteinuria positive cases only [Table/Fig-3,4]. Age ranged from five months to 83 years, where 38.4% were <14 years and 61.6% were >14 years of age. Male to female ratio was found to be 1:1.5. Among these 32 culture positive cases, 62.5% patients presented with sign and symptoms of Urinary tract infections (20/32) while 37.5% were asymptomatic (12/32). There was no significant association between all the urinary parameters (pyuria, culture positivity) and age, sex, and symptoms of UTI among proteinuria positive cases. Association of culture positivity with proteinuria as well as pyuria was found to be statistically significant (p<0.001 and p=0.0353 respectively) [Table/Fig-5]. The positive predictive value of proteinuria was 43.8% while, negative predictive value was 87.8%.
Characteristics of proteinuria positive cases (n=73).
| Characteristics | Percentage (%) |
|---|
| Age |
| <14 years | 28 (38.4) |
| >14 years | 45 (61.6) |
| Sex |
| Male | 29 (39.7) |
| Female | 44 (60.3) |
| Pyuria |
| Present | 31 (42.5) |
| Absent | 42 (57.5) |
| Culture |
| Positive | 32 (43.8) |
| Negative | 41 (56.2) |
Documentation of pyuria and culture by age, sex and symptoms among proteinuria positive cases (n=73).
| Parameters | Pyuria | p-value | Culture | p-value |
|---|
| + | - | + | - |
|---|
| Age (years) |
| <14 | 9 | 17 | 0.335(NS) | 10 | 18 | 0.335(NS) |
| >14 | 22 | 25 | 22 | 23 |
| Sex |
| Male | 16 | 12 | 0.054(NS) | 12 | 16 | 0.8875(NS) |
| Female | 15 | 30 | 20 | 25 |
| Symptoms |
| Symptomatic | 28 | 20 | 0.087(NS) | 20 | 28 | 0.627(NS) |
| Asymptomatic | 9 | 16 | 12 | 13 |
Fisher’s-exact test
Association between UTI and other urinanalysis parameters among proteinuria positive cases (n=73).
| Parameters | Culture result | p-value |
|---|
| + | - |
|---|
| Proteinuria | + | 32 | 41 | <0.001 (S) |
| - | 36 | 260 |
| Pyuria | + | 18 | 13 | 0.0353 (S) |
| - | 14 | 28 |
Fisher’s-exact test
Out of 296 proteinuria negative cases, 36 were found to be culture positive showing significant growth. Antimicrobial resistance of isolates was compared among proteinuria positive (32) and negative cases (36) [Table/Fig-6]. β-lactam antibiotic resistance among proteinuria positive cases and chloramphenicol resistance among proteinuria negative cases was significantly high (p=0.001 and p=0.0100 respectively).
Comparison between urine culture isolates and their antimicrobial resistance profile from proteinuria positive (n=32) and proteinuria negative (n=36) cases.
| Variables | Total (% resistance) | p-value |
|---|
| Proteinuria (+) (n=32) | Proteinuria (-) (n=36) |
|---|
| Antimicrobials |
| β-lactams | 68.7 | 0 | 0.001 (S) |
| Aminoglycosides | 18.5 | 10 | 0.1528 (NS) |
| Cephalosporins | 48.1 | 55 | 0.3960 (NS) |
| Fluoroquinolones | 25.9 | 35 | 0.2190 (NS) |
| Carbapenems | 0 | 0 | 1.000 (NS) |
| Chloramphenicol | 3.7 | 15 | 0.0140 (S) |
| Urinary isolates |
| Escherichia coli | 65.6 | 56.6 | 0.2449 (NS) |
| Klebsiella spp. | 6.2 | 21.8 | 0.0018 (S) |
| Enterococcus | 9.4 | 4.3 | 0.2507 (NS) |
| Citrobacter | 6.2 | 0 | 0.0289 (S) |
| Proteus mirabilis | 3.2 | 0 | 0.2462 (NS) |
| Pseudomonas spp. | 0 | 4.3 | 0.1212 (NS) |
| Acinetobacter spp. | 0 | 4.3 | 0.1212 (NS) |
| Coagulase negative staphylococci | 6.2 | 8.7 | 0.5928 (NS) |
| Candida spp. | 3.2 | 0 | 0.2462 (NS) |
Fisher’s-exact test
The percentage resistance of other antibiotic groups was not significant. Klebsiella spp. were more commonly isolated from proteinuria negative cases (p=0.0018) while Citrobacter spp. from proteinuria positive cases (p=0.0289) [Table/Fig-6].
Discussion
Many clinicians just request for routine microscopic examination of urine without culture and rely on urinalysis parameters as indicator of probable UTI and guide their treatment based on the positive results. In this study, proteinuria was evaluated as predictor of UTI. Although quantitative urine culture is the gold standard for diagnosis of UTI [7]. Several rapid tests are used to rule out UTI for the time and cost effective processing of urine samples received in the laboratory [8]. In our laboratory the cost of urine routine and microscopy is approximately 10 INR per specimen processed, while cost per test of urine culture is approximately 150 INR. Thus, urine culture comes out to be about 15 times more costly compared to urine routine and microscopic examination. Dipstick urinalysis along with pyuria and bacteriuria is the most widely used cost effective method for diagnosis of UTI [9]. Most of the studies have analysed leucocyte esterase and nitrite test in the assessment of UTI [10], proteinuria as a specific parameter for UTI has not been studied much. Proteinuria has been reported as an important parameter that differentiates acute pyelonephritis from other UTIs [3]. In a study conducted in 2014 prevelance of proteinuria in acute phase of pyelonephritis was found to be between 90.9%-98.7% [3].
We established a significant association between culture positivity and proteinuria as well as pyuria (p<0.001, p=0.0353 respectively). UTI can have varied clinical presentation ranging from dysuria, urgency and frequency among adults [2]; and fever, vomiting, diarrhoea, jaundice, failure to thrive, lethargy and frequency of micturition in neonates and young children [11]. In our study, 65.8% study subjects presented with sign and symptoms of UTI. Among the proteinuria positive cases, gender, age and symptoms of UTI had no significant association with pyuria. In a study conducted among school children in Spain, no association was found between asymptomatic UTI and proteinuria, while urine protein levels increase during symptomatic UTI [12]. In another study, the prevalence of proteinuria during bacteruria was 5%-9% [13]. The positive predictive value of proteinuria in our study was 43.8%. Another study has evaluated the positive predictive value of leucocyte esterase and nitrite test (68%) but not proteinuria [7]. However, the negative predictive value of proteinuria in our study came out to be 87.8% which implies that if there is no proteinuria then the probability of UTI is low.
Among the urinary isolates, Klebsiella spp. was found to be significantly associated with proteinuria negative cases as compared to proteinuria positive cases (p=0.0018). This could be related to the production of urease enzyme by Klebsiella spp.,Citrobacter spp. was found to be significantly associated with proteinuria (p=0.0289). While resistance to beta-lactam antibiotics was also significantly high among proteinuria positive cases (p=0.001). On literature search we could not find the association of these parameters with proteinuria in any study. Since culture results are obtained after 24-48 hours, awareness of the presence of proteinuria in a urine sample may help in initiating empirical treatment of UTI.
Limitation
In present study, sample size was limited. May be a similar study on a larger scale will establish the relationship of proteinuria with uropathogens and their antimicrobial resistance pattern; and guide empirical antibiotic prescription for UTI among proteinuria positive cases.
Conclusion
Based on the present study we conclude, proteinuria is not a sufficiently strong indicator of UTI as a single parameter but may have a good predictive power when combined with the other urinalysis parameters and clinical presentation in the diagnosis of UTI. Since culture results are obtained after 24-48 hours, awareness of the presence of proteinuria in a urine sample may help in initiating empirical treatment of UTI. Due to the lack of research on the predictive power of proteinuria for UTI, the relationship between proteinuria and UTI has not been well established. More similar studies are recommended in the future to confirm the predictive role of proteinuria in UTI.
Fisher’s-exact test
Fisher’s-exact test
Fisher’s-exact test