Materials and Methods
This prospective period study was conducted at Sri Ramachandra Medical College and Research Institute after obtaining Institutional ethics committee approval. The study was conducted form July 2015 to June 2016.
Inclusion Criteria
All isolates of Salmonella from clinical samples with a relevant clinical history.
Exclusion Criteria
Duplicate isolates from the same patient were excluded.
Bacterial Isolates
One hundred and three Salmonella isolates from clinical specimens such as blood (94), stool (7), cerebrospinal fluid (1) and pus from splenic abscess (1) collected were included in the study. All the isolates were identified up to species level by conventional /automated methods using VITEK-2 system (Vitek2 GN-card; BioMerieux, Brussels, Belgium). The isolates were serotyped on the basis of agglutination with somatic O, phase I flagellar H antigens by the slide agglutination tests with antisera (King Institute, Chennai) as specified by the Kauffmann- White scheme.
Antimicrobial Susceptibility Testing
By the disk diffusion antimicrobial susceptibility was determined on Mueller-Hinton agar and interpreted according to the guidelines of the CLSI 2015. The antibiotics tested were ciprofloxacin (5 μg), ceftriaxone (30 μg) and cefotaxime (30 μg), chloramphenicol (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), ampicillin (10 μg), azithromycin (15 μg). MIC to cefotaxime and ceftriaxone was determined by agar dilution method with the range tested being 0.008-128 μg/ml and interpreted in accordance with CLSI guidelines 2015 [10].
Detection of the ESBL phenotype was performed by the combined disk method using cefotaxime (30 μg) and ceftazidime (30 μg), based on the inhibitory effect of clavulanic acid [5,7]. Escherichia coli (ATCC® 25992™) (negative control) and Klebsiella pneumoniae (ATCC® 700603™) (positive control) were used as controls. The zone diameter difference of >5 mm for either antimicrobial agent tested in combination with clavulanic acid versus the zone diameter of the agent when tested alone, was considered indicative of production of ESBL
Polymerase Chain Reaction (PCR) amplification
All isolates were subjected to PCR using group specific primers to characterise the blaCTX-M [10]. Co-existence of qnrA, qnrB and qnrS were also looked for [11]. The primers used in the study are shown in [Table/Fig-1].
Primers used in the study.
| Gene | Primer Sequence 5′- 3′ | Product size |
|---|
| blaCTX-M | CTX-M-F: CGCTGTTGTTAGGAAGTGTGCTX -M-R: GGCTGGGTGAAGTAAGTGAC | 754bp |
| qnrA | qnrA- F: ATTTCTCACGCCAGGATTTGqnrA- R: GATCGGCAAAGGTTAGGTCA | 516-bp |
| qnrB | qnrB- F: GATCGTGAAAGCCAGAAAGGqnrB- R: ACGATGCCTGGTAGTTGTCC | 469-bp |
| qnrS | qnrS- F: ACGACATTCGTCAACTGCAAqnrS- R : TAAATTGGCACCCTGTAGGC | 417-bp |
F- Forward primer.
R- Reverse primer.
Extraction of DNA: DNA was extracted from the study isolates by boiling method. A single colony was inoculated into 1.5ml of Luria Bertani broth and incubated overnight. This was centrifuged at 10,000 rpm for 10 minutes. The pellets were suspended in 500μl of distilled water and lysed by heating at 100°C for 10 minutes and centrifuged for 1 minute. The supernatant was utilized as a template for amplification.
PCR conditions for blaCTX-M: 2 μl of the supernatant was mixed in the 23 μl of the master mix, which contained 0.1 μl of Taq polymerase (Takara Bio Inc.) in 2.5 μl of 10x Taq polymerase buffer, 0.5 μl of dNTP ((Takara Bio Inc.), 1 μl of primer and 18.9 μl Milli Q. Amplification reactions were performed under the following conditions: initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds with an extension at 72°C for 50 seconds, and a final extension for one cycle at 72°C for 5 minutes. The PCR product was then run on a 1.5% agarose gel for detection of the amplified fragment [11].
PCR conditions for qnrA, qnrB and qnrS: A 2 μl of the supernatant was mixed in the 23 μl of the master mix, which contained 0.1 μ l of Taq polymerase (TaKaRa Bio Inc.) in 2.5 μl of 10x Taq polymerase buffer, 0.5 μl of dNTP (TaKaRa Bio Inc.), 1 μl of primer and 18.9 μl Miili Q.
Amplification reactions were performed under the following conditions: initial denaturation at 94°C for 5 minutes, followed by 32 cycles of denaturation at 94°C for 45 seconds, annealing at 53°C for 45 seconds with an extension at 72°C for 60 seconds, and a final extension for one cycle at 72°C for 5 minutes. The PCR product was then run on a 1.5% agarose gel for detection of the amplified fragment [12].
DNA Sequencing
The DNA of blaCTX-M positive isolates was extracted with PureLink Genomic DNA Kit (Invitrogen), according to the user manual provided with the kit and subjected to automated DNA sequencing (ABI 3100, Genetic Analyser, Applied Biosystems, Foster city, CA, USA). The aligned sequences were analysed with the Bioedit sequence program and similarities searches for the nucleotide sequences were performed with the BLAST program.
Clinical History
The clinical history of the patients with ESBL producing Salmonella infection was collected retrospectively from the medical records department. The data collected were the chief complaints during admission, course in the hospital stay, treatment given and the outcome.
Results
Of the 103 study isolates, the majority were Salmonella typhi (68), followed by Salmonella paratyphi A (26). The others serotypes were Salmonella typhimurium (7) and Salmonella enteritidis (2).
The antimicrobial susceptibility profile of the 103 Salmonella isolates was ampicillin 87.4% (90/103), chloramphenicol 96.1% (99/103), cotrimoxazole 96.1% (99/103) ciprofloxacin 91.2%(94/103), ceftriaxone 98.1%(101/103), cefotaxime 98.1%(101/103) and azithromycin (100%).
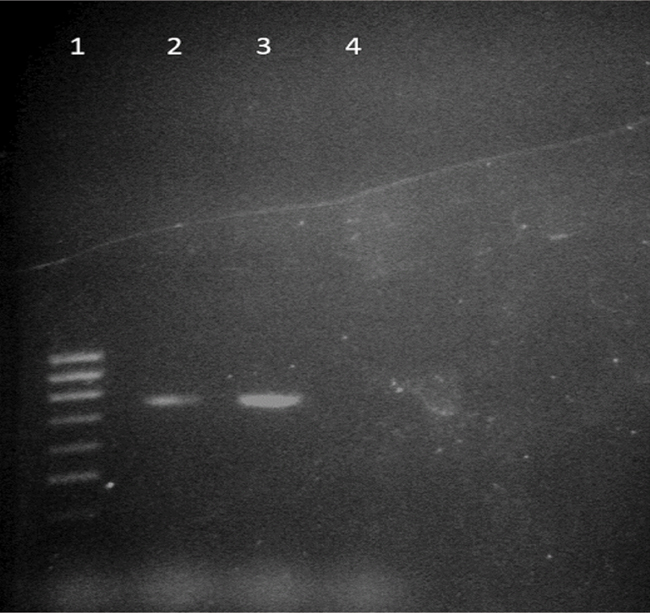
MIC90 to both cefotaxime and ceftriaxone was 1 μg/ml. Two isolates of Salmonella typhi which were isolated from blood culture were resistant to cefotaxime and ceftriaxone and had a MIC of 128μg/ml. PCR amplification and sequencing detected the presence of blaCTX-M-15 in these isolates. PCR for blaCTX-M-15 is shown in [Table/Fig-2]. qnr was not detected in any of the study isolates.
PCR for blaCTX-M 15.
Lanes 1: Molecular mass marker (100bp DNA ladder); Lane 2: Positive control (Previous positive blaCTX-M was confirmed by gene sequencing); Lane 3: blaCTX-M15 positive (amplicon size- 754 bp); Lane 4: Negative control - E.coli ATCC® 25992™
Discussion
Enteric fever is endemic in India with Salmonella enterica var Typhi (S. typhi) and Salmonella enterica var Paratyphi A (S. paratyphi A) being the major causative agents [1]. These pathogens are transmitted by the faeco-oral route in regions with poor standards of hygiene and sanitation accounting for high morbidity and mortality. Antibiotic therapy constitutes the mainstay of management of enteric fever. Failure to treat an infection properly leads to prolonged illness, complications and development of carrier state. Fluoroquinolones and third-generation cephalosporins were used for the treatment of salmonellae infections after the emergence of MDR Salmonellae (resistant to ampicillin, chloramphenicol and co-trimoxazole) [9,13,14]. This lead to the occurrence of reduced susceptibility to ciprofloxacin. Resistance to third-generation cephalosporins among salmonellae though low at present (1%) is also emerging in India [3].
Plasmid-mediated quinolone resistance is mediated by qnr genes which encode a protein that protects DNA gyrase from ciprofloxacin and by aac(6′)-Ib-cr, an aminoglycoside modifying enzyme with activity against ciprofloxacin. Plasmids bearing qnr or aac(6′)-Ib-cr may also carry an extended spectrum cephalosporin resistance gene, which results in the development of resistance to both fluoroquinolone and cephalosporin and thereby limiting the therapeutic options available for the management of invasive salmonellosis [4,15]. In this study the CTX-M bearing salmonellae did not harbor the qnr genes, though they exhibited resistance to ciprofloxacin in vitro. Hence, the presence of other quinolone resistance mediators like gyr A, gyr B and par C should be looked for.
In countries like Germany, Philippines and Kuwait CTX-M-15 and SHV-12 has been described in typhoidal Salmonellae which showed high-level resistance to ceftriaxone. S. typhi producing ACC-1 AmpC β- lactamase has been reported from India [4,16]. Though ESBL producing Non typhoidal Salmonella have been cited worldwide, their occurrence in serovar Typhi and Paratyphi A is not common [9,15,17].
The incidence of CTX-M-15 among enteric fever producing salmonellae is not alarming as evident in this study, but the potential for their widespread dissemination may result in treatment failures. In many tropical countries including the Indian subcontinent, the widespread availability and uncontrolled use of antibiotics can lay the foundation for rapid dissemination [18].
Since, the blaCTX-M is located on plasmids in conjunction with mobile genetic elements such as IS EcpI, they disseminate rapidly. Gene transfer experiments in many studies have been performed to localize their position on plasmids. But non-transferrable blaCTX-M has also been described suggesting their possible chromosomal location. There is a reservoir of ESBLs and CTX-M genes among Escherichia coli and Klebsiella pneumoniae which constitutes a huge risk factor for spread of resistance to other pathogenic Enterobacteriaceae [19]. Additionally, use of third generation cephalosporins to treat enteric fever and other infections caused by Salmonella species may the increase the incidence of CTX-M producing salmonellae. Decreased susceptibility to ciprofloxacin among Salmonella species has also contributed to the increased use and subsequent development of resistance to cephalosporins among salmonellae. The continued surveillance of cephalosporin resistant Salmonella in combination with prudent use of these agents both in animals and humans is crucial for limiting the spread of CTX-M producing salmonellae [19].
There are 2 reports of occurrence of CTX-M15 type ESBL in Salmonella enterica. This includes two Indian patients with Salmonella typhi and 2 others with paratyphi C and paratyphi A. All were travellers returning from India to UAE and Japan respectively [17,20].
The occurrence of blaCTX-M15 in conjunction with quinolone resistance is alarming because it further limits the therapeutic options available for typhoid fever especially in endemic countries such as India [15]. In the past CTX-M15 ESBLs have been found exclusively in Escherichia coli and Klebsiella species. Salmonella strains can acquire the gene encoding for this enzyme from Escherichia coli and Klebsiella species in the community. This speculation is supported by the fact that the mobile genetic element ISEcpI is responsible for mobilization of the bla genes and it is identified upstream of several blaCTX-M genes [17] .
Azithromycin is presently being used for the management of uncomplicated typhoid fever and associated with a prompt resolution of clinical symptoms and low prevalence of relapse and convalescent fecal carriage [12]. However, azithromycin treatment failure in a patient with invasive salmonellae infection has been described [21].
Conclusion
To conclude, enteric fever producing salmonellae harbor the CTX-M gene thus making cefotaxime ineffective for treatment. This also implies that use of other third generation cephalosporins for treatment may result in failure of therapy. Though the incidence of CTX-M in this study is not alarming, the potential for rapid dissemination due to the location of the genetic elements and the huge reservoir encountered in Enterobactericeae is a cause for concern.
Limitation
In present study, sample size was too small to conclude any statistically significant results. Also, further studies on Salmonella harboring TEM, SHV and PER genes needs to be carried out as these genes also confers resistance to third generation cephalosporins.