The mechanisms of antibiotic resistance in enterococci are by mutation and horizontal gene transfer mediated via plasmids and transposons [5]. Glycopeptide resistance is mediated by different Vancomycin resistance (Van) gene operons namely VanA, VanB, VanC, VanD, VanE, VanG, VanL, VanM and VanN. Out of these VanA is most common followed by VanB and VanC is responsible for the intrinsic resistance present in E. gallinarum and E. casseliflavus [6].
Vancomycin predisposes patients to colonization and infection with VRE by inhibiting growth of normal Gram-positive bowel flora and by providing advantage for VRE that may be present in small numbers in an individual’s bowel [4]. The primary sites of colonisation in the hospitalised patients are gastrointestinal tract, skin and soft tissues. The present study was carried out for identification of species, antimicrobial resistance pattern and genotypic characterization of VRE isolates from clinically significant infections in hospitalized patients and their association with gut colonization.
Materials and Methods
This cross-sectional descriptive study was carried out over a period of one and half years during October 2013 to April 2015 at the Department of Microbiology, Vardhman Mahavir Medical College and Safdarjung Hospital, New Delhi, India. A total of 4200 samples (pus, blood and urine) received from hospitalized patients were processed according to standard protocols [7]. Patients positive for enterococci isolates were included in this study. Patients positive for VRE isolates were assessed for gut colonization by inoculating fecal sample onto vancomycin screen agar. The study protocol was approved by the Institutional Ethical Committee (IEC/Nov 13/98).
Inclusion Criteria
Clinically significant enterococci isolated from pus, blood and urine samples from hospitalized patients.
Exclusion Criteria
Enterococci isolates from samples collected from patients presenting at the outpatient department.
A total of 250 non-duplicate clinically significant enterococci isolates were included in the study. The isolates were identified by their colony morphology, catalase reaction, growth on bile esculin agar, tolerance to 6.5% NaCl, growth at 10°C and 45°C. Species identification was carried out by a battery of biochemical tests; carbohydrate fermentation tests (mannitol, arabinose, sucrose, sorbitol, lactose and raffinose), reduction of potassium tellurite, arginine hydrolysis tests, motility and pigment production [8]. E. faecalis and E. faecium were confirmed by PCR using specific primers and controls [Table/Fig-1]. The unidentified species were confirmed by Vitek 2 system (bioMeriux, USA). The antibiotics tested were ampicillin (10 μg), ciprofloxacin (5 μg), high content gentamycin (120 μg), vancomycin (30 μg), teicoplanin (30 μg), linezolid (30 μg), and tigecycline (15 μg). In addition, nitrofurantoin (300 μg) and norfloxacin (10 μg) were included for urinary isolates.
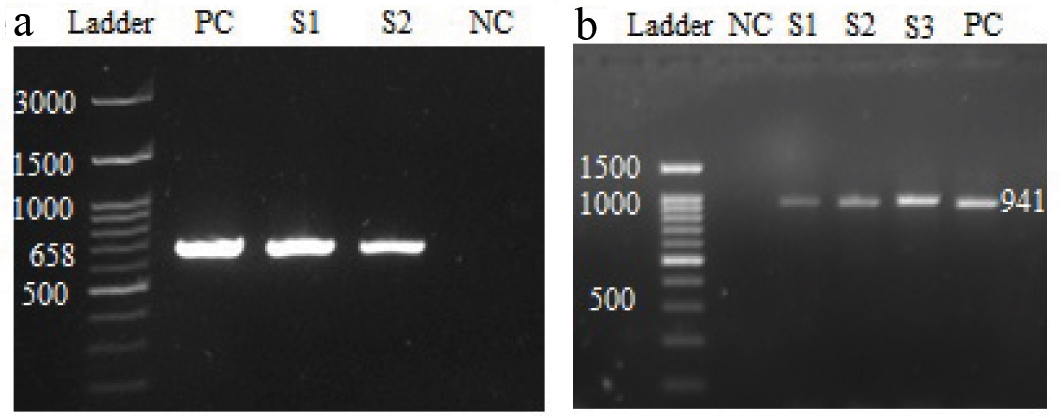
a) Agarose gel electrophoresis of the amplified products by PCR for E. faecium (658 bp). Ladder – 100 bp DNA ladder, S1, S2 – sample; PC- Enterococcus faecium ATCC® 19434 TM was used as positive control; NC- Mili-Q water was used as negative control; b) Agarose gel electrophoresis of the amplified products by PCR for E. faecalis (941 bp). Ladder - 100 bp DNA ladder; S1, S2, S3- sample; PC- Enterococcus faecalis ATCC® 29212 TM was used as positive control; Mili-Q water was used as Negative Control (NC).
Screening for presence of VRE was carried out by agar dilution method. About 10 μL of bacterial inoculums (0.5 McFarland turbidity standards) were spot inoculated onto the Mueller Hinton Agar (MHA) plates with different concentrations (0.25-32 μg/mL) of vancomycin (HiMedia, Mumbai) and incubated at 37°C for 24 hours. Growth of more than one colony was considered positive [9]. Finally, MIC was confirmed by E test (bioMeriux, France).
All isolates with vancomycin MIC ≥ 2 μg/mL were tested against teicoplanin, linezolid, tigecycline, quinupristin-dalfopristin and daptomycin by E-test. Antibiotic susceptibility results were interpreted according to CLSI 2012 guidelines [9]. The European Committee on Antimicrobial Susceptibility Testing guidelines (EUCAST- 2012; www.eucast.org) were used for tigecycline [10].
Genotypic Characterization of Vancomycin Resistance Genes
Strains were inoculated in Luria-Bertani broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl), incubated overnight aerobically at 37°C and DNA was extracted by phenol chloroform method as described by Sambrook J et al., [11].
PCR reaction was performed in a thermocycler (Eppendorf, USA). Briefly initial denaturation at 94°C for five minutes followed by 30 cycles of denaturation at 94°C for one minute, annealing temperature (differs according to the genes) for one minute, amplification at 72°C for one minute for 30 cycles and final extension at 72°C for 10 minutes. PCR assay was carried out in sterile 0.2/0.5 mL PCR tubes with 25 μL reaction volume. Each 25 μL reaction mixture contained 2.5 μL 10X PCR buffer, 2.5 μL 1.5 mmol MgCl2, 2 μL 0.2 mmol dNTPmix, 0.5 μL of 10 pmol each of forward and reverse primers, 0.2 μL Taq DNA polymerase (1 unit/reaction) and sterile double distilled water to make the final volume. Primers used in the study are described in [Table/Fig-2].
Details of primers used in the study.
| Genes | Amplicon size (bp) | Primers |
|---|
| VanA | 732 | FP 5’ GGGAAAACGACAATTGC 3’RP 5’ GTACAATGCGGCCGTTA 3’ |
| VanB | 536 | FP 5’ AAGCTATGCAAGAAGCCATG 3’RP 5’ CCGACAATCAAATCATCCTC 3’ |
| VanC1 | 822 | FP 5’ TGGTATCAAGGAAACCTCGC 3’RP 5’ CCGACTTCCGCCATCATAGCTG 3’ |
| E. faecalis | 941 | FP 5’ ATCAAGTACAGTTAGTCT 3’RP 5’ ACGATTCAAAGCTAACTG 3’ |
| E. faecium | 658 | FP 5’ TTGAGGCAGACCAGATTGACG 3’RP 5’ TATGACAGCGACTCCGATTCC 3’ |
| cfr | 746 | FP 5’ TGAAGTATAAAGCAGCTTGGGAG 3’RP 5’ ACCATATAATTGACCACAAGCAG 3’ |
FP – Forward Primer, RP – Reverse Primer, bp- Base Pairs
The primers were sourced from Eurofins Genomics, Bangalore. The quality control strains used were Staphylococcus aureus ATCC® 25923™, E. faecalis ATCC® 29212™, E. faecalis ATCC® 51299™, E. faecium ATCC® 700221™, E. faecium ATCC® 19434™. Mili-Q water was used as negative control. (GenBank Accession number of VanA, VanB, VanC1-E. faecalis and E. faecium genes are CP019995.1, U00456.1, EU151771.1, CP003726.1 and CP003583.1 respectively).
Multiplex PCR (Eppendorf, USA) was carried out to detect the presence of genes encoding for vancomycin resistance. Of the many genotypes of vancomycin resistance described in enterococci, attempt was made to identify the most common ones, i.e., VanA, VanB and VanC genotypes [Table/Fig-3a,b]. The linezolid resistant E. faecium was tested for presence of cfr gene by PCR [Table/Fig-4].
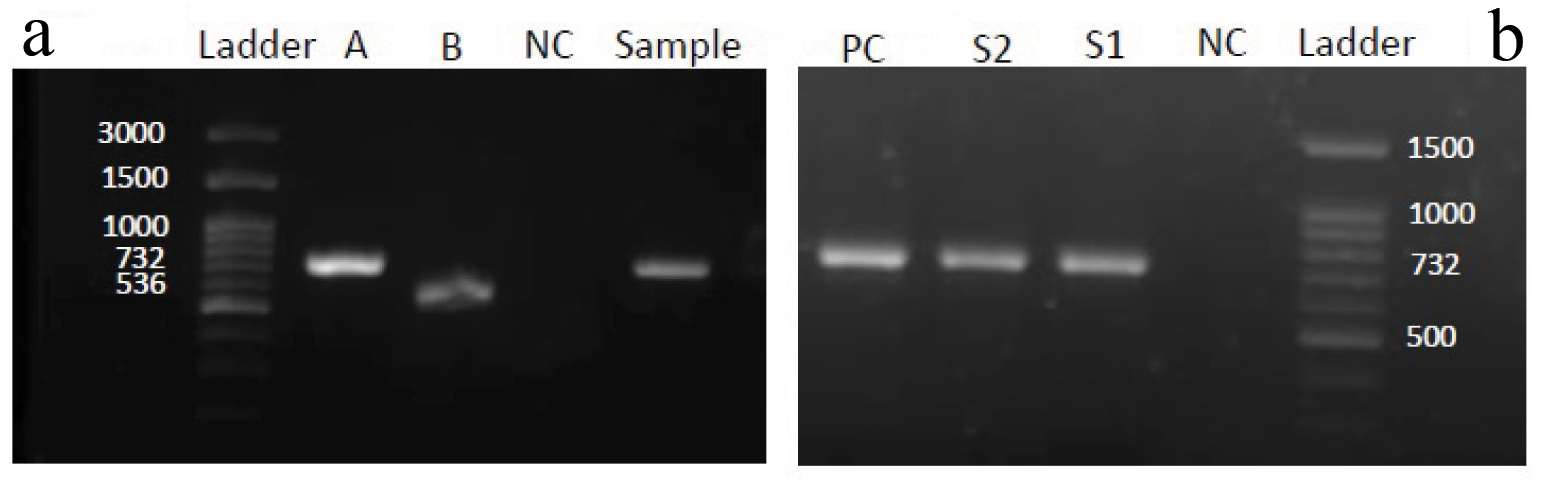
a) Agarose gel electrophoresis of the amplified products by PCR for vanA(732 bp) and VanB(536 bp) genes. Ladder 100bp DNA ladder, A and B - positive control for vanA (E. faecium ATCC®700221TM) and VanB (E. faecalis ATCC® 51299TM), NC- Mili-Q water was used as negative control; b) Agarose gel electrophoresis of the amplified products by PCR for vanA(732 bp). Ladder 100bpDNA ladder,S1- clinical sample positive for vanA and S2 – faecal sample from same patient positive for vanA, PC-E. faecium ATCC®700221TM was used as positive control for vanA; NC- Mili-Q water was used as negative control.
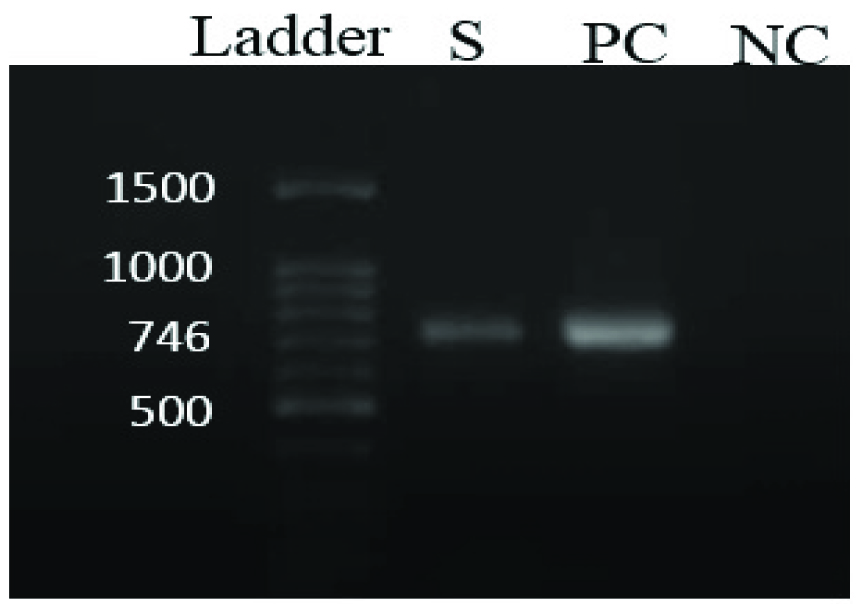
Agarose gel electrophoresis of the amplified products by PCR for cfr gene(746 bp).
Ladder- 100bp DNA ladder, S- sample, PC- Staphylococcus aureusATCC® 25923TM was used as positive control, NC- Mili-Q water was used as negative control
After completion of PCR reaction, 10 μL of each PCR product were mixed with 2 μL of bromophenol blue dye. This mixture was loaded in well of 1.5% agarose gel and electrophoresed at constant 80 volts with Tris -Acetate Ethylene Diamine Tetra Acetic acid (TAE) buffer. Molecular marker of 100 bp DNA ladder was run concurrently in a separate well. The gel stained with 0.5 mg/mL of ethidium bromide was visualised using a UV transilluminator (Alpha Innotech, Cell Biosciences, USA).
Clinical history of patients infected with VRE isolates was obtained and were screened for gut colonization with VRE. Fecal samples were inoculated onto vancomycin screen agar with 6 μg/mL of vancomycin, incubated overnight at 37°C. VRE isolates were further processed for species identification and genotypic characterization.
Statistical Analysis
SPSS software version 21.0 was used. Continuous data was analysed using the Student’s t-test. Categorical data was assessed using Pearson’s Chi-square test. A value of p ≤ 0.05 was considered statistically significant.
Results
Out of 4200 samples processed, a total of 250 (5.9%) enterococci were isolated. The isolates were from urine (128; 51.2%), blood (75; 30.0%), and pus (47; 18.8%). The distribution of enterococci isolates in hospitalized patients is described in [Table/Fig-5]. The isolates were from 182 (72.8%) adults and 68 (27.2%) paediatric patients. Male to female ratio in adult age group was 1:1.2 and in paediatric age group it was 1:1.
Distribution of enterococci and VRE isolates among different clinical departments. Figures in parenthesis are percentages.
| Location | Enterococci isolatesN=250(%) | VRE isolatesN=63(%) |
|---|
| Paediatric ward | 66 (26.4) | 17 (27.0) |
| ICU[Adult (47), Paediatric (2)] | 49 (19.6) | 27 (42.8) |
| Obstrtrics and Gynaecology | 45 (18.0) | 7 (11.1) |
| Medicine | 36 (14.4) | 2 (3.2) |
| Nephrology | 23 (9.2) | 7 (11.1) |
| Orthopaedics | 15 (6.0) | 1 (1.6) |
| Surgery | 11 (4.4) | 1 (1.6) |
| Burns ICU | 5 (2.0) | 1 (1.6) |
Species Distribution among the Enterococci Isolates
E. faecium (162, 64.8%) was the most common species followed by E. faecalis (82, 32.84%). Motile enterococci (6, 2.4%), were confirmed as E. gallinarum by Vitek 2 system (bioMeriux, USA).
Antimicrobial Resistance among Enterococci Isolates
Among 250 enterococci isolates, 174 (69.6%) were resistant to ampicillin, 187 (74.8%) to high level gentamycin, 240 (96.0%) to ciprofloxacin, 57 (22.8%) to vancomycin and 56 (22.4%) to teicoplanin by disk diffusion method. Among urinary isolates, 42 (32.8%) and 118 (92.2%) isolates showed in vitro resistance to nitrofurantoin and norfloxacin respectively. One isolate was also resistant to linezolid. Although all isolates were susceptible to tigecycline by disk diffusion method, discordant results were observed in two isolates, which were resistant with MIC near breakpoints (MIC= 1.5 and 2μg/mL). E. faecium was more resistant species as compared to E. faecalis [Table/Fig-6].
Antibiotic resistance pattern of enterococci isolates by disc diffusion method. Figures in parenthesis are percentages.
| No. of Isolates (%) | % Resistance |
|---|
| Gentamycin(120 μg) | Ciprofloxacin(5 μg) | Ampicillin(10 μg) | Vancomycin(30 μg) | Teicoplanin(30 μg) | Nitrofurantoin^(300 μg) | Norfloxacin^(10 μg) |
|---|
| E. faeciumN=162 (64.8) | 135 (83.3) | 162 (100) | 156 (96.3) | 55 (33.9) | 54 (33.3) | 36/84 (42.8) | 84/84 (100) |
| E. faecalisN=82(32.8) | 52 (63.4) | 74 (90.2) | 18 (21.9) | 2 (2.4) | 2 (2.4) | 6/43(13.9) | 34/43 (79.1) |
| *E. gallinarumN=6 (2.4) | 0 | 4 (66.7) | 0 | 0* | 0 | 0 | 0 |
| TotalN=250 | 187 (74.8) | 240 (96.0) | 174 (69.6) | 57 (22.8) | 56 (22.4) | 42/128 (32.8) | 118/128 (92.2) |
*E. gallinarum strains are intrinsically resistant to vancomycin, but they may appear sensitive by disc diffusion method.
^ Tested only in urinary isolates.
There was a good correlation between vancomycin agar dilution and E test. Vancomycin MIC ranged 0.25-1 μg/mL in majority of isolates (187, 74.8%). Six (2.4%) isolates MIC range was 2-3 μg/mL and were identified as E. gallinarum. High level vancomycin resistance was observed in rest 57 (22.8%) isolates (MIC= 64 ->256 μg/mL). Among isolates with high level vancomycin resistance (n= 57), 56 isolates were resistant to teicoplanin (MIC = 32 - >256 μg/mL) suggestive of VanA phenotype. One isolate was sensitive to teicoplanin (MIC = 8μg/mL) suggestive of VanB phenotype. The six E. gallinarum isolates were sensitive to teicoplanin (MIC from 0.5-1 μg/mL).
Majority (57/63, 90.5%) of VRE isolate carried VanA gene. Species distribution among the VanA gene carriers was E. faecium (55, 87.3%) followed by E. faecalis (2, 3.2%). Rest (6, 9.5%) VRE isolates were of E. gallinarum species carried VanC1 gene. VanB genotype was absent in the present study. The phenotypic VanB isolate was identified as VanA genotypically.
Gut colonization could be studied only in 47/63 patients with VRE infections. Majority 33/47 (70.2%) were colonized with VRE. The respective fecal isolates of enterococci were phenotypically (species, antibiogram) and genotypically (resistance gene) similar to the clinical isolates [Table/Fig-3b]. Risk factors for VRE gut colonization were evaluated and only the sex of the patients was significantly associated (p=0.045) [Table/Fig-7].
Demographic data and risk factors associated with VRE gut colonization. Student’s t-test and Pearson’s Chi-square test were used for analysing continuous and categorical data respectively.
| Variable | Class | Gut colonization | p-value* |
|---|
| Positive (33) | Negative (14) |
|---|
| Sex | Male | 20 (60.6%) | 4 (28.6%) | 0.045 |
| Female | 13 (39.4%) | 10 (71.4%) |
| Age group | Adult | 27 (81.8%) | 11 (78.6%) | 0.796 |
| Paediatric | 6 (18.2%) | 3 (21.4%) |
| Metronidazole inj. | Yes | 12 (36.4%) | 7 (50.0%) | 0.384 |
| No | 21 (63.6%) | 7 (50.0%) |
| Vancomycin | Yes | 8 (24.2%) | 3 (21.4%) | 0.835 |
| No | 25 (75.8%) | 11 (78.6%) |
| Intravenous catheter | Yes | 32 (97.0%) | 13 (92.8%) | 0.512 |
| No | 1 (3.0%) | 1 (7.2%) |
| Dialysis | Yes | 11 (33.3%) | 3 (21.4%) | 0.503 |
| No | 22 (66.7%) | 11 (78.6%) |
| Nasogastric tube | Yes | 21 (63.6%) | 9 (64.3%) | 0.966 |
| No | 12 (36.4%) | 5 (35.7%) |
| Tracheostomy | Yes | 3 (9.1%) | 2 (14.3%) | 0.627 |
| No | 30 (90.9%) | 12 (85.7%) |
| Ventilator use | Yes | 17 (51.5%) | 6 (42.8%) | 0.587 |
| No | 16 (48.5%) | 8 (57.2%) |
| Length of hospitalization (days) | Mean (Range) | 15.73 (5-130) | 12.71 (5-20) | 0.6 |
*p value ≤ 0.05 is significant.
The genetic characteristics of VRE and MIC50, MIC90, MIC range of vancomycin, teicoplanin, daptomycin, linezolid, tigecycline and quinupristin-dalfopristin are shown in [Table/Fig-8]. All the VRE isolates were susceptible to daptomycin (MIC = 0.094–4 μg/mL), quinupristin-dalfopristin (MIC = 0.023-1 μg/mL). Two (3.2%) isolates were resistant to tigecycline (MIC = 1.5 and 2 μg/mL). One (1.6%) of the VRE isolate was resistant to linezolid (MIC = 12 μg/mL). cfr gene was detected in a linezolid resistant isolate by PCR.
Phenotypic and genotypic characterization of VRE isolates with MICs for various antibiotics. No VanB genotype detected.
| Phenotype | Genotype | Species | MIC of higher antibiotics (μg/mL) |
|---|
| Vancomycin | Teicoplanin | Daptomycin | Linezolid | Tigecycline | Quinupristin-Dalfopristin |
|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range |
|---|
| VanA(n=56)VanB(n=1) | vanA(n=57) | E.faecium(n=55)E.faecalis(n=2) | >256 | >256 | 64-512 | 24 | 64 | 8-512 | 2 | 4 | 0.094-4 | 0.75 | 2 | 0.047-12 | 0.064 | 0.25 | 0.023- 2 | 0.064 | .25 | 0.023-1 |
| VanC(n=6) | vanC1(n=6) | E.gallinarum(n=6) | 2 | 3 | 2-3 | 0.25 | 1 | 0.125-1 | 0.125 | 2 | 0.064-2 | 0.064 | 0.125 | 0.047-0.125 | 0.064 | 0.125 | 0.023-0.125 | 0.047 | 0.25 | 0.023-0.25 |
Discussion
Vancomycin resistant enterococci have been reported from many countries of the world since 1986 [2]. Widespread use of vancomycin and extended-spectrum cephalosporins in hospitals contributed to the worldwide emergence of VRE and has made it difficult to treat serious enterococcal infections over the past 20 years [12].
The prevalence of VRE infections in India is also increasing in the past one decade. Mathur P et al., from New Delhi was the first to report VRE from India in 1999 [13]. The reports of prevalence of VRE from India vary from 1% to 8.7%. In the present study, a total of 63 (25.2%) VRE isolates were obtained and rate is high as compared to other Indian studies [3,14-17]. Out of 63 VRE strains, 32 (50.8%) were from urine, 25 (39.7%) from blood, and 6 (9.5 %) from pus samples. In this study, majority of VRE isolates were from ICU [Table/Fig-5], and the finding is similar to another study from Northern India [3].
Glycopeptide resistance as per literature is mediated by six different Van gene operons namely VanA, VanB, VanC, VanD, VanE and VanG. Recently new gene clusters have been discovered (VanL, VanM, and VanN). In the present study, most (57, 90.5%) of the VRE isolates carried VanA gene suggesting high level resistance to glycopeptides. Rest (6, 9.5%) were positive for VanC1 gene suggesting intrinsic low level resistance to vancomycin. One isolate was VanB phenotype-VanA genotype and in concurrence with another Indian study [17].
Among the various antimicrobials evaluated for treatment of serious infections with VRE are quinupristin-dalfopristin, linezolid, daptomycin and tigecycline. In the present study, one VRE isolate was resistant to linezolid (MIC=12 μg/mL) and two isolates were resistant to tigecycline (MIC = 1.5 and 2 μg/mL). On genotypic analysis, cfr gene was detected in the linezolid resistant strain. Linezolid resistant enterococci have been recently reported from India [18]. In another study cfr gene has been detected in linezolid resistant staphylococci [19]. Yasliani S et al., reported two isolates of E. faecium resistant to linezolid from Tehran with MIC 32 μg/mL [20]. Linezolid resistance has increased substantially after inclusion of linezolid on the hospital antibiotic policy [21]. Tigecycline resistance in VRE isolates has been reported from Taiwan and Turkey [22,23]. In India, tigecycline resistance in VRE isolates has not been reported so far.
There are several studies for the surveillance of colonization of Enterococcus spp. in the gastrointestinal tract, a major site of initial colonization. In a surveillance study from two ICUs in Brazil, VRE strains were isolated from 32.6% patients in rectal swabs and in 20% VRE isolated in their clinical samples also [24]. In a study from ICU and post-surgery ward in a hospital in Iran, 29.3% patients had rectal colonization for VRE [25]. In the present study, among patients with infection due to VRE, gut colonization was observed in 33/47 (70.2%). Detection of phenotypically and genotypically (VanA) similar isolates from clinical sample and gut is indicative of endogenous infection. Gut colonization with VRE may be a source of endogenous infection or horizontal transfer among hospitalized patients due to breach in infection control practices. Based on the high colonization status, review of the ongoing antibiotic policy and active VRE surveillance as an integral part of infection control strategy were suggested [25]. To the best of our knowledge, detection of cfr gene in linezolid resistant E. faecium and tigecycline resistance in VRE isolate along with characterization of VRE and their association with gut is reported for the first time from India.
Limitation
Gut colonization hypothesis could not be confirmed by molecular studies like Pulse Field Gel Electrophoresis (PFGE).
Conclusion
Vancomycin resistance among enterococci is an emerging threat. Emergence of tigecycline and linezolid resistance mediated by cfr gene further narrows therapeutic options. Gut colonization with VRE may be a source of endogenous infection and horizontal transmission within the hospital. The threat of untreatable enterococcal infection and the possibility that vancomycin resistance may spread from enterococci to other common Gram-positive bacteria namely staphylococci argue for strict surveillance.