Enterococci are Gram positive cocci, known only as intestinal commensals with little significance. These have evolved as deadly pathogens over last two decades. Enterococcal infection is always difficult to treat due to its inherent resistance to many commonly used antibiotics like β-lactams, aminoglycosides (when used alone), cephalosporins, co-tromoxazole and clindamycin [1,2]. The mainstay of the treatment of enterococcal infection over the years was penicillin with gentamycin due to their synergistic action. By 1979, resistance to high-level gentamycin was reported due to genetically acquired mechanisms which can be by mutation or through acquisition of DNA. Today acquired resistance has rendered many of the circulating strains of enterococci resistant to other available therapeutic options as well [1-4]. Presently many circulating strains are reported to have acquired resistance to most of the remaining therapeutic options including vancomycin and linezolid which are thought to be antibiotics of last resort in enterococcal infection [4]. Enterococci are usually associated with hospital acquired infections like urinary tract infections, surgical site infections, bacteraemia, endocarditis, and rarely meningitis [1-4].
In USA alone, vancomycin resistant enterococci associated healthcare infection amounts to ten thousand per year [4]. Enterococci are considered as the second leading cause of hospital acquired infection [4-8]. The overall national data of enterococcal infection rate is yet to be available from India due to paucity of cumulative data collection system. Hence, the present study was aimed at detecting the antimicrobial resistance pattern among Enterococcus isolates obtained from clinical specimens at a tertiary care centre in Western Uttar Pradesh, with a special emphasis on aminoglycoside, vancomycin and linezolid resistance and to discuss the therapeutic option on those multidrug resistant enterococcal isolates.
Materials and Methods
A total of 15342 clinical samples (urine, blood, pus, genital swab, others) were collected from patients suspected of bacterial infection visiting the IPD/OPD of Subharti Medical College, Meerut between May 2014 to April 2015. All the samples were processed in the Microbiology department using standard microbiological techniques for isolation and identification. All samples except blood were cultured on blood agar, chocolate agar and MacConkey’s agar plate and incubated at 37°C for 24 and 48 hours. The plates were observed for growths which were identified using standard bacteriological guidelines [2]. Blood received for culture was processed in BacT/ALERT® 3D system (Biomerieux) and sub cultured on above mentioned solid media after positive flagging.
Out of 15342 samples, 5370 (35%) were culture positive. The Enterococcus species were identified upto species level by Gram stain, catalase test, 6.5% NaCl tolerance test, heat tolerance test, bile esculin test and a set of biochemical tests i.e., arginine, arabinose, sucrose, sorbitol, mannitol, raffinose and pyruvate [2]. The culture media was procured from Hi-media Laboratories Pvt. Ltd., Mumbai, India.
Inclusion criteria- only samples with positive growth of Enterococcus were included. Exclusion criteria- isolates that were commensals/colonizers were excluded. Among the culture positive samples; 242 (4.5%) were identified as Enterococcus species. On clinical follow up, 42 isolates were excluded from the study as they were found to be a colonizer or were commensal and only the remaining 200 non repeat isolates were subjected for further identification and sensitivity.
The approval from the Institutional Research and Ethical committee was obtained before conducting the study.
Antibiotic susceptibility testing was carried out for the isolates by Kirby-Bauer disk diffusion method on Mueller Hinton agar as per CLSI 2014 guidelines [9], using commercially available antibiotic discs (HiMedia, Mumbai, India). The antibiotics tested for various microorganisms and their disc potency are as follows: penicillin (10 U), ampicillin (10 μg), ciprofloxacin (5 μg), erythromycin (15 μg), doxycycline (30 μg), tetracycline (30 μg), linezolid (30 μg); and for urine isolates nitrofurantoin (300 μg) and norfloxacin (10 μg) were used.
For detecting High-Level Gentamycin (HLG) and High-Level Streptomycin (HLS) resistance three methods were used: (i) disc diffusion using gentamycin (120 μg) and streptomycin (300 μg) dics; (ii) Agar dilution method; and (iii) Broth microdilution as per CLSI 2014 guidelines [9].
Vancomycin resistance was screened by disc diffusion method uising vancomycin (30 μg), teicoplanin (30 μg). Vancomycin resistant isolates were further confirmed by agar dilution method and by determining MIC with Vitek 2 (Biomerieux) [9].
In absence of molecular facility the vancomycin resistant enterococci were classified as Van A and Van B on the basis of relative MIC of vancomycin and teicoplanin owing to the fact that strains of Van B phenotype remain susceptible to teicoplanin; whereas, Van A phenotype is resistant to both the glycopeptides (Van A: teicoplanin MIC ≥16 μg/mL, Van B teicoplanin MIC ≤0.5 μg/mL); and also to the fact that Van A and VanB are two most common circulating phenotype of VRE among E. faecalis, and E. faecium [10,11].
Linezolid resistance detected in disc diffusion test was also confirmed by MIC breakpoints using CLSI 2014 guidelines [9]. Quality control was done using Enterococcus faecalis ATCC® 29212™.
Statistical Analysis
Statistical analysis was done using Z proportion test and SPSS-16.0 statistical software (IBM) and p-values less than 0.05 were considered as statistically significant.
Results
Out of 200 isolates of Enterococcus studied, 169 (84.5%) were E. faecalis, 27 (13.5%) were E. faecium and 4 (2%) were E. casseliflavus. Enterococci were isolated predominantly from IPD patients (189, 94.5%) than from OPD patients (11, 5.5%). Maximum number of isolates were from female patients (142, 71%) as compared to male patients (58, 29%). The age distribution ranged from 5 day to 86 years with maximum number of isolates in the age group 20 to 29 years (47, 23.5%). Alarmingly, 22 (11%) isolates were from the age group 0-10 years and 4 (2%) isolates were each from neonates (0 to 28 days) and infants (28 days to one-year-old) [Table/Fig-1]. Urine was the most common sample (140, 70%); followed by pus (29, 14.5%), blood (14, 7%) and genital swabs (9, 4.5%) [Table/Fig-2].
Age wise distribution of patients from which Enterococcus is isolated with respect to OPD/IPD (N=200).
| Age | Number | OPD | IPD |
|---|
| 0-28 days | 4 | 0 | 4 |
| 28 days -1 year | 4 | 0 | 4 |
| >1-10 years | 14 | 0 | 14 |
| 11-19 years | 9 | 0 | 9 |
| 20-29 years | 47 | 6 | 41 |
| 30-39 years | 31 | 3 | 28 |
| 40-49 years | 31 | 1 | 30 |
| 50-59 years | 34 | 1 | 33 |
| 60-69 Years | 14 | 0 | 14 |
| 70-79 Years | 9 | 0 | 9 |
| 80-89 years | 3 | 0 | 3 |
| Total | 200 | 11 | 189 |
Speciation and sample distribution of Enterococcus isolates (N=200).
| S.NO | SAMPLE | E. faecalis (%) | E. faecium (%) | E. casselif-lavus (%) |
|---|
| 1. | Urine (140) | 127(90.7%) | 10(7.14%) | 3(2.14%) |
| 2. | Pus (29) | 20(68.9%) | 8(27.5%) | 1(3.4%) |
| 3. | Blood (14) | 13(92.8%) | 1(7.14%) | - |
| 4. | Genital swab 9) | 3(33.3%) | 6(66.6%) | - |
| 5. | Others (8) | 6 (75%) | 2(25%) | - |
| TOTAL | 200 | 169(84.5%) | 27(13.5%) | 4(2%) |
Among all the enterococcal isolates; 25 (12.5%) were HLGR, 13 (6.5%) were HLSR and 62 (31%) were HLGR and HLSR [Table/Fig-3].
Showing sensitivity to high-level aminoglycosides among enterococcal.
| Variables | E. faecalis | E. faecium | E.casseliflavus | Total |
|---|
| HLGR | 21 | 3 | 1 | 25(12.5%) |
| HLSR | 10 | 3 | 0 | 13(6.5%) |
| HLGR+HLSR | 47 | 14 | 1 | 62(31%) |
| Sensitive to High-level Gentamicin & Streptomycin | 91 | 7 | 2 | 100(50%) |
*HLGR=High-Level Gentamycin Resistance, HLSR: High-Level Streptomycin Resistance.
The frequency of high-level aminoglycoside resistance (HLGR, HLSR and both) was found more in E. faecium as compared to E. faecalis, which was statistically significant (p<0.05 by Z proportion test) [Table/Fig-4].
Comparison of HLGR, HLSR HLAR found among enterococcus faecalis and E. faecium.
| SPECIES | HLGR | HLSR | HLGR +HLSR |
|---|
| Enterococcus faecalis (169) | 40% | 33.7% | 27.8% |
| Enterococcus faecium (27) | 62.9% | 62.2% | 51.8% |
| p value | 0.0257 | 0.004 | 0.0124 |
Highest resistance was seen with erythromycin (168, 84%), followed by ciprofloxacin (164, 82%); and penicillin, ampicillin (151, 75.5% each). However, doxycycline (93, 46.5%) and tetracycline (95, 47.5%) were shown to have moderate resistance against Enterococcus. In urine samples, norfloxacin resistance was seen in 17 (83.5%) isolates and nitrofurantoin resistance was seen in 46 (32.85%) isolates [Table/Fig-5].
Showing antimicrobial drug resistance in Enterococcus isolates.
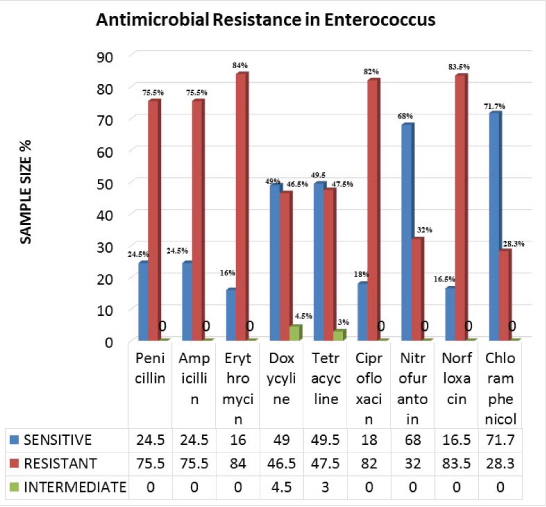
Vancomycin resistance was seen in 14 (7%) isolates and all the isolates were E. faecalis. The 14 VRE isolated, 11 (78.5%) were Van A and 3 (21.4%) were Van B phenotypes. Sample wise distribution and MIC of Van A and Van B are described in [Table/Fig-6].
Samples and speciation of VRE isolated with respect to MIC of vancomycin and teicoplanin all vancomycin resistant isolates were from indoor patients.
| Variables | No. of Isolates | Sample | Species | Vancomycin MIC(μg/mL) | Teicoplanin MIC(μg/mL) |
|---|
| ≥ 32 | ≥ 64 | ≤ 8 | ≥ 32 |
|---|
| Van A | 11 | Urine - 7Blood-3Endo* - 1 | E. faecalis | 9 | 2 | - | 11 |
| Van B | 3 | Urine -3 | E. faecalis | 3 | - | 3 | - |
*Endocervical swab
Out of 14 VRE isolates, 12 (85.7%) isolates were resistant to both HLG and HLS. The VRE isolated were resistant to most of the antibiotics; however, some sensitivity was seen in ciprofloxacin (28.5%) doxycycline (92.8%), ampicillin (14.2%) and nitrofurantoin (30%) as described in [Table/Fig-7].
Sensitivity pattern of VRE isolated.
| Antibiotics | Total | Sensitive | Percentage |
|---|
| Penicillin and Ampicillin | 14 | 2 | 14.2% |
| Erythromycin | 14 | 3 | 21.4% |
| Doxycycline | 14 | 13 | 92.8% |
| Tetracycline | 14 | 12 | 85.7% |
| Ciprofloxacin | 14 | 4 | 28.57% |
| Norfloxacin | 10 | 0 | 0% |
| Nitrofurantoin* | 10 | 3 | 30% |
| Chloramphenicol** | 4 | 3 | 75% |
| HLS# | 14 | 2 | 14.2% |
| HLG## | 14 | 2 | 14.2% |
| Linezolid | 14 | 8 | 57.4% |
*used only in urinary isolates;
** Not reported in urine samples, HLS#: High-Level Streptomycin, HLS: High-Level Gentamycin
Linezolid resistance was seen in 4 (2%) E. feacalis isolates with MIC ≥8 μg/mL, isolated from clinical samples (three urine samples, one blood sample) of IPD patients.
These four linezolid resistant isolates were also VRE, HLGR and HLSR. However, these isolates were sensitive to doxycycline (3/4, 75%), tetracycline (2/4, 50%), and nitrofurantoin (2/3, 66.6%).
Discussion
In present study, out of 200 Enterococcus isolates studied; 189 (94.5%) isolates were obtained from IPD, were clearly in excess of 11 (5.5%) isolates obtained from OPD. Similarly HLAR, VRE, linezolid resistant isolates were also frequent in IPD samples as compared to OPD samples indicating that most of these infections are of hospital origin. Similiar findings have also been reported in literature [1-7]. Maximum numbers of Enterococcus isolates were from urine samples (140, 70%). Similar finding of higher rate of enterococci isolated from urine samples have been reported by Kanthishree BH et al., 72.2% [12] and by Chakraborty A et al., 66% [13]. The reason for high rate of isolation of enterococci from females (71%) in this study might be due to the proximity of urethra to the perineal area due to shorter length of urethra in females leading to higher rate of UTI in females. The second most common sample in this study was pus (29, 14.5%) as also reported by Kanthishree BH et al., 16.6% [12] and by Sharma R et al., 22% [14] in their study.
HLAR was found to be more in E. faecium (51.8%) than E. faecalis (27.8 %). Similar findings were seen by Mendiratta DK et al., and by Fernandes SC et al., [15,16]. Fernandes SC et al., showed HLAR in 35.7% of E. faecalis and 35.3% of E. faecium [16]. In this study HLGR, HLSR and their mix presence (HLGR+HLSR) was found to be higher in E. faecium than in E. faecalis (p-value of 0.0257, 0.004 and 0.0124, respectively).
The rate of VRE was 7% in the present study. Similar findings of VRE have been reported in studies by Fernandes SC et al., 8.6% [16]. Praharaj I et al., 8.7% [17] and Ghazawy IF et al., 6.3% [18] Praharaj I et al., showed prevalence of Van A and Van B as 90.6% and 6.25% respectively, which was similar to present study [17]. As compared to a study done by Tripathi A et al., isolation rate of VRE was 7.9% and all were of Van A phenotype [19]. In contrast Phukan C et al., found VRE to be 24%, this might be due to different selection criteria as that study was done only on 67 isolates [20].
The rates of VRE and linezolid resistance reported from similar studies are shown in [Table/Fig-8] [17-21].
Comparison of prevalence of VRE and linezolid resistance in different studies.
| S.NO | Author | Place and Year | VRE | Linezolid Resistance |
|---|
| Percentage | Van A | VanB |
|---|
| 1. | Praharaj I et al [17] | Puducherry, India 2013 | 8.7% | 90.6% | 6.25% | 0% |
| 2. | El-Ghazawy IF et al [18] | Egypt, 2016 | 6.3% | 100% | 0% | 2.1% |
| 3. | Tripathi A et all [19] | Lukhnow, India 2016 | 7.9% | 100% | 0 | 0% |
| 4. | Phukan C et al [20] | Guwahati, India 2016 | 24% | 56.25 | | 4.5% |
| 5. | Sundaram M et all [21] | Karaikal, India 2016 | 0 | - | - | 0% |
| 6. | Present study | Meerut, India | 7% | 78.5 | 21.4% | 2% |
Emergence of resistance against the antibiotics like vancomycin or linezolid which was considered as last resort till now, is a matter of concern which emphasizes the need to explore other therapeutic options in such conditions.
Glycopeptides like teicoplanin are effective against VRE that expresses Van B phenotype than Van A phenotype. However, in those strains expressing Van B phenotype, development of resistance to teicoplanin has been noted [22].
Lower urinary tract infections due to VRE can be treated with nitrofurantoin that achieves adequate level in urine but not in blood [22]. In present study, 10 (71.4%) of 14 VRE isolates were isolated from urine samples and sensitivity to nitrofurantoin was 30%. This indicates the importance of culture and sensitivity in all critical cases which will help in treating infection with VRE with a cheaper and easily available drug like nitrofurantoin in at least 30% of VRE associated UTIs as seen in present study. Other alternatives like fosfomycin can also be used [22].
There has been some interest in the use of quinolones against VRE. While ciprofloxacin typically displays bacteriostatic activity against enterococci, synergic bactericidal activity has been demonstrated in vitro when combined with ampicillin or gentamycin against susceptible strains [23]. In our study, overall ciprofloxacin sensitivity was 18% and 28.5% in VRE and overall doxycycline sensitivity was 46.5% and 92.8% in VRE.
Linezolid an oxazolidinone, introduced only in year 2000, exerts antibacterial activity by inhibiting the formation of the 70S initiation complex becomes the drug of choice for many type of VRE. Linezolid resistance in Enterococcus was reported from India in 2013 [17,24] and since then there has been increase in incidence of reporting linezolid resistant VRE [17,19-21].
A major advantage of linezolid is the availability of both parental and oral formulations and the oral formulation is almost 100% bioavailable [22]. In this study, linezolid resistance was 2%; similar results were seen in other studies as shown in [Table/Fig-8] [17,19-21].
All the linezolid resistant isolates were E. faecalis which was VRE as well as HLAR which is a major therapeutic concern. Mutation in the genes encoding 23S rRNA, an important part of drug binding site of ribosome is the most common mechanism of linezolid resistance and this selection for mutated genes in rRNA was originally demonstrated in staphylococci and have subsequently identified in enterococci as well, and it is associated with longer duration of therapy.
This calls for proper use and de-escalation whenever linezolid is intended for use. The other mechanism of linezolid resistance transferable plasmid-mediated resistance to linezolid due to cfr gene is a major threat due to its potential to spread across species [25]. In these cases of concurrent linezolid and vancomycin resistance treatment option includes available agents which do not have a specific VRE approval (chloramphenicol, doxycycline, high dose ampicillin or ampicillin/salbactum) [26]. For high dose ampicillin or ampicillin/salbactum MIC and β-lactmase production needs to be done routinely for all isolates which is yet a dream in all laboratories in a resource poor country like India.
Newer therapeutic option like quinupristin/dalfopristin and daptomycin has greatly increased the therapeutic option for the treatment of serious VRE. Quinupristin/dalfopristin, a combination of Sreptogramin A (dalfopristin) and streptogramin B (quinupristin) and daptomycin a lipopeptide, antimicrobial agent is effective for VRE. But quinupristin and dalfopristin have proved to be intrinsically resistant in case of E. faecalis hence effective only against E. faecium [22]. Unfortunately most of the VRE isolated in hospital set up are E. faecalis which consist of all the VRE or linezolid resistant Enterococcus in present study.
The peptide antibiotic daptomycin is rapidly bactericidal against VRE but initial studies have shown, it is poorly effective in vivo [23]. Oritavancin, a new semi synthetic glycopeptide has demonstrated in vitro activity against VRE. In August 2014, the United States FDA approved oritavancin for treatment of skin infections only [27]. Tigecycline is a derivative of minocyclines that has activity against VRE in vitro. But these drugs are yet to pass the initial hurdles and their sensitivity guideline is yet to be defined in CLSI in enterococcal isolate. As most of these multidrug resistant enterococcal infections are hospital acquired strict adherence to preventive measures like adherence hand hygiene, strict monitoring, reporting and isolation and barrier nursing of subjects from where these MDR organism are isolated remains the only way to prevent such infections.
Limitation
Due to limitation of resource and cost, molecular confirmation of VRE phenotypes was not done in this study. However, initial presumption of Van A and Van B types by MIC to vancomycin and teicoplanin help us in treating the patients with limited resource as teicoplanin is a therapeutic option in Van B phenotypes. Also, this characterization helps to reduce the cost burden, in selection of primers, when molecular typing is done.
Conclusion
The emergence of 2% linezolid resistant VRE from the hospital is a matter of concern as till now they are considered to be last resort for treatment in patients infected with VRE. Very few reports of linezolid resistant VRE are reported from India so it has become a cryptic problem because to the best of our knowledge there is no published data from this part of Western Uttar Pradesh leading to lack of awareness and indiscriminate use of linezolid. The approved drug in such case quinupristin/dalfopristin is inherently resistant to most commonly isolated enterococcal species (E. faecalis). This calls for the need of strict enforcement of antibiotic policies, coupled with greater adherence to infection control measures to prevent emergence and spread of antibiotic resistant bacteria.
*HLGR=High-Level Gentamycin Resistance, HLSR: High-Level Streptomycin Resistance.
*Endocervical swab
*used only in urinary isolates;
** Not reported in urine samples, HLS#: High-Level Streptomycin, HLS: High-Level Gentamycin