To predict the clinical outcomes of the infection and also for better understanding the distribution of microorganism and its evolutionary origins, it is very important to study the diversities of H. pylori genes. In the present study, we tried to explore the distribution of various genotypes of H. pylori isolates from biopsy specimens and also to establish the potential association with the clinical outcome.
Materials and Methods
This prospective cross-sectional study was conducted at the Department of Microbiology, Shree P.M. Patel college of Paramedical Science and Technology, Anand, Gujarat, for a period of one year from October 2012 to September 2013.
Design of the Study
Seventy one consecutive (46 males and 25 females, age; 10-90 years), symptomatic patients attending the endoscopic unit of “deep surgical hospital” were included in this study. Based on endoscopic findings, out of 71 patients, 34 patients were suffering from gastritis, 26 with reflux oesophagitis, nine with duodenal ulcer and two with duodenitis. Patients taking aspirin or Non-Steroidal Anti-Inflammatory Drugs (NSAIDS) in the past four week or those on Proton Pump Inhibitors (PPI) or patients with previous therapy to eradicate H. pylori, or if the inform consent was not obtained were excluded from the study.
Sample
One antral biopsy was collected from each symptomatic patient in fasting condition using Olympus Video Endoscope 1306.
Ethical Considerations
The present study got ethical approval from Human Research Ethics Committee (HREC) of H.M. Patel Center for Medical Care and Education, Pramukh Swami Medical College, Karamsad, Gujarat, India.
DNA Extraction from Biopsy Sample
DNA isolation from 71 biopsy specimens was done by using “QIAamp DNA mini kit” (Qiagen GmbH, Hilden, Germany, Lot No: 11872534, Cat No: 51306) as described by the manufacturer.
Genomic DNA Isolation
The tissue biopsies were centrifuged at 5000 rpm for 10 min and re-suspended in 200 μL of ATL (Animal Tissue Lysis) buffer for complete lysis. Finally, the DNA was eluted in 100 μL of elution buffer. DNA purity and quantity was determined using a GeneSys 10UV spectrophotometer (Thermo Scientific, USA). The extracted DNA was stored at -20°C until used as template in PCR.
H. pylori Genotyping
PCR reaction was carried out in a final volume of 50 μL as follows. 5 μL of 1X PCR buffer (50 mM KCL, 10 mM tris HCL, 2.5 mM MgCl2, 0.01% gelatin), 8 μL of 0.2 mM dNTPs, 0.5 μL each primer (Procured from Bangalore Genie), 0.6 μL of Taq DNA polymerase, 6 μL DNA sample and 25.4 μL distilled water. Thermal cycler (ESCO-2720) of Applied Biosystem was used for amplification. [Table/Fig-1] summarizes the primer sequences, expected size of the product and PCR conditions. The amplified PCR products were analysed by agarose gel electrophoresis. An 8μL of PCR product were run on a 2.5% agarose gel, containing ethidium bromide and were visualized under ultraviolet light source.
PCR primers and conditions.
| Gene | Primer sequence | PCR conditions | Size of the PCR product (bp) | Reference |
|---|
| cagA | F-5′-GATAACAGCC AAGCTTTTGAGG-3′R-3′-CTGCAAAAGA TTGTTTGGCAGA-5′ | denaturation for 1 min at 94°C; annealing for 1 min at 56°C and extension for 1 min at 72°C (35 cycles) | 349 | [15] |
| ureC | F-5′-AAGCTTTTAGG GGTGTTAGGGGTT-3′R-3′-AAGCTTACTTT CTAACACTAACGC-5′ | denaturation for 1 minute at 93 °C; annealing for 1 minute at 55 °C and extension for 1 minute at 72 °C (35 cycles) | 294 | [15] |
| vacA | F-5′-ATGGAAATA CAACAAACACAC-3′R- 3′-CTGCTTGA ATGCGCCAAAC-5′ | denaturation for 1 min at 94°C; annealing for 1 min at 53°C and extension for 1 min at 72°C (35 cycles) | 286 | [15] |
| iceA | F-5′-GTGTTTTTAA CCAAAGTATC-3′R-3′-CTATAGCCA TTATCTTTGCA-5′ | denaturation for 1 min at 94°C; annealing for 1 min at 56°C and extension for 1 min at 72°C (35 cycles) | 247 | [15] |
F: Forward primer; R: Reverse primer; bp: Base pairs
Statistical Analysis
The Fischer’s-exact and Chi square (χ2) test were used to compare the relationship between H. pylori genotypes and clinical outcome. A p-value of < 0.05 was considered statistically significant.
Results
Detection of the Three Genes by PCR
Out of the 71 biopsies screened, 22 (31%) samples were positive for H. pylori by PCR, with high proportion of cagA positive (17/22 specimen; 77.27%), followed by ureC positive (4/ 22 specimen; 18.18%) and vacA positive (1/22 specimen; 4.54%) strains [Table/Fig-2]. we did not find any strain with iceA gene in our province. Forty nine (69%) biopsy samples were negative for all the four genes screened.
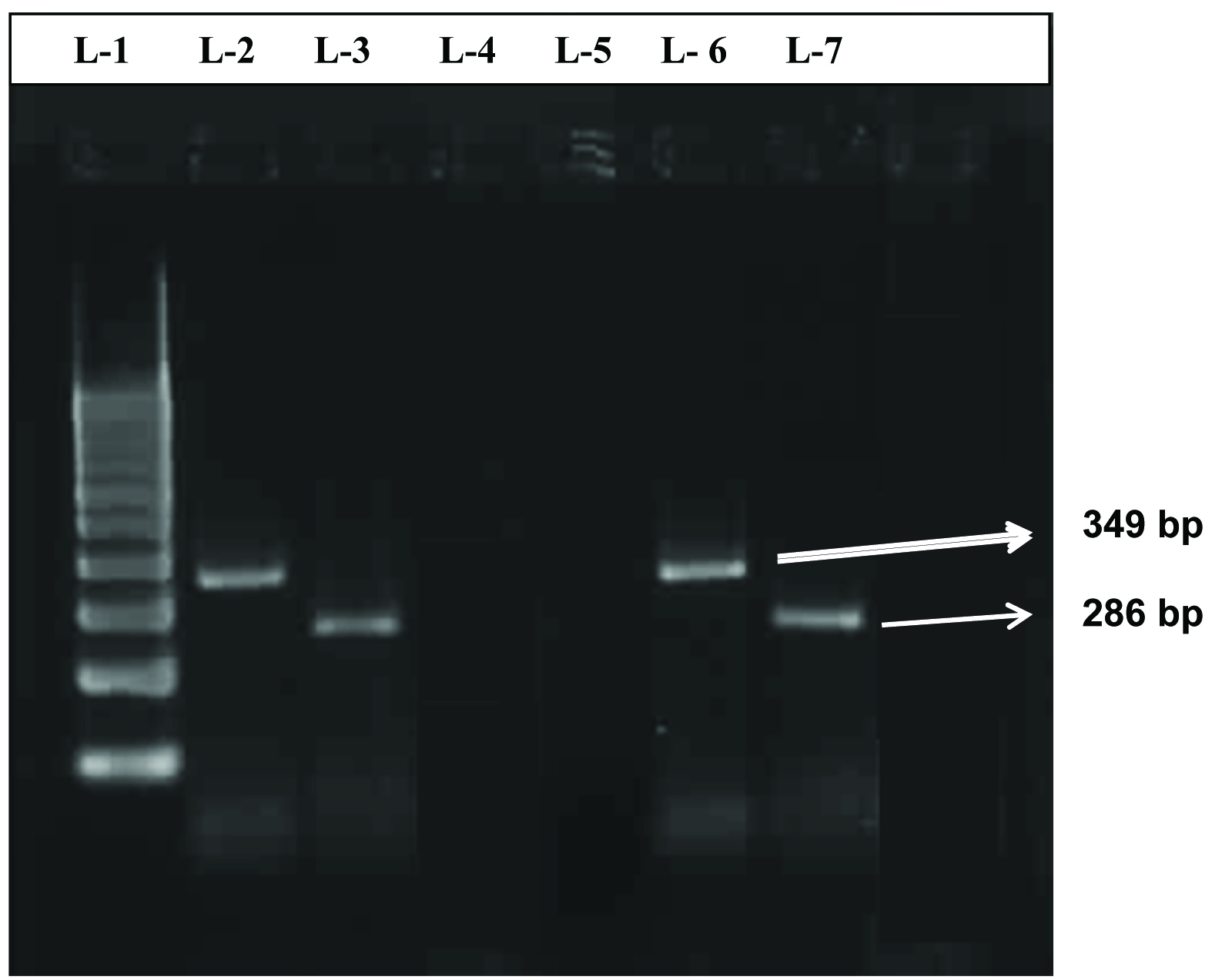
Separation of PCR products by 2.5% agarose gel electrophoresis.
Legend for figure: Lane 1: 100bp DNA ladder, Lane 2 and 3: cagA positive control and vacA positive control, lane 6: cagA gene (349bp), Lane 7: vacA gene (286bp), Lane 4: negative control.
Association of cagA gene with various demographic factors like age group and gender as well as with gastrointestinal diseases
Prevalence of cagA was found higher in age group 21-40 years. (52.94%), followed by 41-60 years (35.29%) and 61-80 years. (11.76%). High incidence of cagA positive H. pylori strains found in adults, may suggest that this population may be at risk for developing more serious pathology of the gastric mucosa. We found significant association between cagA and gender of the patient; prevalence was higher in females (10/17, 58.8%), as compared to males (7/17, 41.2%) (p-value 0.042 < 0.05) [Table/Fig-3]. We found that out of 17 cagA positive strains, 9(52.94%) were found in patients with gastritis, 5(29.41%) in reflux oesophagitis and 3(17.64%) in patients with diodenal ulcer [Table/Fig-4].
Distribution of H. pylori genotypes in different gender.
| Genes | Male | Female | chi-square(p-value) |
|---|
| cag A | 7 | 10 | 4.10 (0.042)* |
| vac A | 1 | 0 | N.A. |
| ice A | 0 | 0 | N.A. |
| ure C | 2 | 2 | 0.258(0.612) |
Chi square (χ2) test, *p-value < 0.05 is significant.
Prevalence of H. pylori cagA, vacA, ure C genotypes in various endoscopic findings.
| Genotype | Duodenal Ulcer(n=9) | Duodenitis(n=2) | Gastritis(n=34) | Reflux oesophagitis(n=26) |
|---|
| cagA (17+ve) | Negative | 6 | 2 | 25 | 21 |
| Positive | 3 | 0 | 9 | 5 |
| Chi-square(p-value) | 0.499(0.480) | 0.648(0.421) | 0.229(0.632) | 0.500(0.479) |
| vacA (1+ve) | Negative | 9 | 2 | 34 | 25 |
| Positive | 0 | 0 | 0 | 1 |
| Chi-square(p-value) | 0.147(0.701) | 0.029(0.864) | 0.932(0.334) | 1.755(0.185) |
| ureC (4+ve) | Negative | 9 | 2 | 32 | 24 |
| Positive | 0 | 0 | 2 | 2 |
| Chi-square(p-value) | 0.615(0.433) | 0.123(0.726) | 0.008(0.931) | 0.327(0.567) |
Detection of ureC gene and vacA and its correlation with various demographic factors like age group, gender and Gastro-intestinal disease
The ureC gene (four strains positive) was found to be equally distributed (50%) in both the gender and age-group of 21-40 and 41-60 years. Prevalence of ureC genotype was 50% in gastritis patient as well as in reflux oesophagitis patients [Table/Fig-4]. We found only one strain positive for vacA in a male patients of reflux oesophagitis belonging to age group of 61-80 years.
Discussion
H. pylori possess a remarkable degree of genetic diversity, which is closely related with its epidemiological and pathological characteristics, as a result genotyping becomes very important to characterize the strains and even if many infections are clinically silent, but the patient infected with H. pylori presents increased morbidity and mortality [10,11]. The present study was designed to analyse the genetic distribution of various genes of H. pylori from biopsy samples and to associate its role in the clinical outcome of infections. We found that prevalence of cagA was 77.27% (17/22), which was in conformity to various studies done where the prevalence varies from 62%-77% respectively [12,13], while in one study done by Udhayakumar G et al., prevalence of cagA positive strain was 96% [14]. The prevalence of cagA genotypes in Anand district is relatively important to reveal the circulating H. pylori strains in a given geographic region and the associated need for region-specific diagnostics for H. pylori virulence markers. The present study revealed a relatively high prevalence of the more virulent allele cagA suggesting a common prevalence of virulent H. pylori genotypes. The cagA positive H. pylori was detected in 52.94% of gastritis patients (p-value, 0.632) and 17.64% of duodenal ulcer patients (p-value, 0.480), statistically the correlation is not found significant, but 9 of 22 cagA positive strains shows emergence of this promising gene in Anand district in relation with the patients suffering from acute gastritis. Results are quite in accordance with many studies done, which proves its predominance in causing gastritis [15,16]. We also found significant association between cagA and female gender (p-value, 0.042), which is in contrast to the study done by Abadi ATB et al., who proved that the presence or absence of the cagA gene is not significantly linked to the gender [17].
H. pylori and Gastroesophageal Reflux Disease (GERD): An Unsolved Mystery
Studies have found a negative association between the presence of positive cagA and gastroesophageal reflux diseases [18]. The relationship between cagA positive H. pylori and complications of GERD is still controversial and the role of virulence markers of the bacterium has not been evaluated in most studies of GERD. In our study, we have attended 26 reflux oesophagitis patients, out of which eight patients were positive for H. pylori infection and out of which, 5 (19.23%) were cagA positive strains, 2 (7.69%) were ureC strains and 1 (3.84%) vacA positive strain [Table/Fig-4]. From the above results, we cannot remark on any protective association between them as majority of H. pylori infected GERD patients were having cagA genotype.
Detection of iceA Gene in Gastric Specimen and in their Corresponding H. pylori Isolates
In our study, we have 0% prevalence of this gene, conversely we had three peptic ulcer patients and all the three were cagA positive but iceA negative. In contrast to our results, a study done by Peek RM et al., showed an association between the allelic variant iceA1 and more severe inflammation of gastric mucosa [19]. Similar studies showing no association between iceA gene and peptic ulcer have been reported from Turkey and Lithuania [20,21]. We were unable to confirm an association between the iceA gene and clinical outcome. This finding may reflect important geographic differences between H. pylori and patients. As it is a well known fact that H. pylori genotypes are not uniformly distributed all over the world.
To the best of our information, this is the first study done in Gujarat state of Western India to find the clinical relevance of putative virulence-associated genes of H. pylori in patients with gastric diseases.
Limitation
The most important constraint of the study is the sample size; large studies with more number of samples are required to prove the role of these markers in virulence.
Conclusion
The virulence strains with cagA genotype extend at our province, may result in severe clinical outcomes such as ulcers which may be developed to cancer. We also draw focus on the 0% prevalence of iceA gene in peptic ulcer patients, which concludes that iceA genotype is not a useful marker of virulence in this population and that the progression from gastritis to peptic ulcer must require some other genes or factors including the genetic susceptibility of the host.