In Egypt, among other skin diseases, the incidence of PV ranged between (10-14%) [12]. Other researchers reported that the incidence of PV increased markedly to about 40.8% and 30% among other dermatomycosis in Assiut Governorate and new valley governorate respectively [13]. Inspite of treatment, the recurrence rate of Malassezia is about 60% in the first year and 80% in the second year. So, accurate laboratory diagnosis of the Malassezia species causing PV disease is required [14,15].
So far, there is still debate on the specific types associated with the development of PV, with varying results from different countries. Little data about Malassezia species is available from Egypt [16,17].
Therefore, the focus of the present study was to identify and analyse the distribution and clinico-epidemiological features of the Malassezia species in both Egyptian PV patients and healthy individuals by using of phenotypic and molecular methods.
Materials and Methods
The study was conducted at outpatient clinics of Dermatology Department of Mansoura University Hospitals from October 2012 to September 2013. The principles outlined in the Declaration of Helsinki were followed and informed consents were obtained from all patients and healthy subjects [18]. The study protocol was approved by the local Ethics Committee of Mansoura University, Egypt.
Patients: A total of 137 clinically suspected PV patients (70 male and 67 female with the mean age 25.5±8.3 years) were included. Their characteristic lesions were scaly hypopigmented or hyperpigmented well-defined macules, with slight desquamation and colour ranging from white to brown [12]. Thirty healthy individuals of matched age and sex (21 male and 9 female with the mean age 21.5±9.2 years) were included as a control group. The samples were taken mainly from healthy skin of chest and back [10].
Inclusion criteria: Characteristic lesions of hypopigmented or hyperpigmented scaly macules and patches which may be associated with itching. The lesions lasting atleast one year were defined as longstanding forms and considered extensive if the achromic and/or hyperchromic lesions were distributed on at least four out of the following nine body sites (face, neck, back, chest, abdomen, axilla, arm, buttocks and genital area) and each site covered by more than 20 lesions [19]. The skin scales were collected where most of the lesions were evident with repeated scraping after asking patients not to wash themselves in the area of hypochromic lesions.
Exclusion criteria: Patients with skin lesions of extensive desquamation or inflammation that did not match PV. Patients who had received topical antifungal therapy within last three months or oral antifungal therapy within the last six months.
Specimen Collection: The lesions of PV patients were first cleaned with 70% ethyl alcohol prior to sampling. Skin scrapings were taken from most scaly site using sterile scalpel blade and transported in a sterile filter paper to the laboratory where further investigations were done. In addition, skin samples from healthy individuals were taken by means of sellotape from different healthy skin areas of chest and back.
Sample Processing
Direct microscopy: Microscopic examination of the scales was performed after treatment with 10-20% KOH. Characteristic spaghetti and meatballs appearance (short curved hyphae and round yeasts) confirmed the diagnosis of PV.
Culture [20,21]: All samples were inoculated in slants of Sabouraud’s Dextrose Agar (SDA) and Sabouraud’s Dextrose over laid with sterile olive oil (Oxoid) for isolation of lipid independent species and lipid dependent species, respectively and supplemented with cycloheximide and chloramphenicol. All the slants were incubated at 32°C and examined on days 3, 7 and then at weekly intervals up to three weeks for any developing colonies.
Smears from the colonies were stained with Gram’s stain and examined under microscope for identification of Malassezia. Mixed cultures were excluded. After three weeks of incubation, the culture slants without growth were considered negative and discarded. In order to achieve pure cultures, for each positive sample five colonies were subcultured on SDA and stored at -20oC until PCR analysis.
Species identification procedures by phenotypic methods [7,21]: Further species identification of Malassezia isolates was done according to their morphological features, physiological properties and conventional mycological methods. They include cultural characteristics, Gram staining for yeast phase, lactophenol cotton blue for mycelial phase and biochemical criteria as catalase test, urease test and Tweens assimilation test (Tween 20, 40, 60, and 80).
Morphological features [14]:Malassezia species were differentiated based on examination of colonial and microscopic morphologies i.e., colonies’ colour, shape and texture on SDA medium, and cell shape, size and bud pattern in stained smears.
Biochemical tests [7,21]: The isolates of M. restricta were identified by the negative catalase test while, the isolates of M. furfur and M. pachydermatis were identified by the positive urease reaction. Moreover, the isolates of M. pachydermatis were identified by ability to grow on a lipid-free medium.
Tween assimilation test [22]: As reported by Kaneko T et al., some Malassezia species have the ability to utilize the different Tween compounds as a unique lipid supplement. From each isolate, a colony suspension (of at least 107cfu/mL) in 2 mL sterilized distilled water was made and poured onto SDA plate. The inoculums were then spread evenly. After solidification of each plate at 45°C, four wells were made and filled with 5 μL of a Tween compound (20, 40 and 80, respectively). These plates were incubated at 32°C for a week and the resulting growth was assessed around each well three times (after 2, 4 and 7 days).
Species Identification By Molecular Methods
DNA extraction: Pure colonies obtained from SDA subculture were subjected to repeated freezing and thawing to dissolve their cell walls mechanically. The QIAamp tissue kit (Qiagen, Germany) was used for DNA extraction from yeast according to manufacturer instructions.
PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) of 26S rDNA [23,24]: PCR-based RFLP technique using restriction enzyme digestion specific for the differentiation of the known 11 Malassezia species was used (M. furfur, M. sympodialis, M. globosa, M. restricta, M. slooffiae, M. obtusa, M. dermatis, M. japonica, M. yamatoensis, M. pachydermatis and M. nana).
For amplification of the highly conserved 26S rDNA region, specific primers were used. Their sequences were as following: forward, 5’-TAACAAGGAT-TCCCCTAGTA-3’ and reverse, 5’-ATTAC-GCCAGCATCCTAAG-3’. Conventional PCR was performed in a 50 μL reaction volume using 25 μL master mix (Fermentas, Life Sciences) containing 25 mM of each deoxynucleoside triphosphate (equimolar concentration of dATP, dCTP, dGTP and dTTP), 1.25 U Taq DNA polymerase and 10x PCR buffer in addition to 2.5 μL each primer, 15 μL distilled water and 5 μL DNA template.
The amplification was performed in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems) and included initial denaturation step at 94°C for 5 minute, followed by 30 cycles of denaturation at 94°C for 45 seconds, annealing at 55°C for 45 seconds, and extension at 72°C for 1 minute, with a final extension step of 72°C for 7 minute. The expected size of the amplified PCR products were of 580 bp and visualized by1.5% (w/v) agarose gel electrophoresis in Tris Borate EDTA (TBE) buffer, stained with ethidium bromide (0.5 μg/mL), in comparison with DNA size marker (Thermo Scientific™ GeneRuler™ 50bp DNA Ladder, USA) and examined under UV trans-illumination.
The restriction enzymes Hha1 and BstC1 (New England Biolabs, Hitchin, UK) were selected to obtain the best species-specific pattern. Digestion was performed by incubating 17 μL aliquot of PCR products with 10 U of the enzyme which sum up to 20 μL. After the reaction at 37°C for three hours, the electrophoresis was done as above and the restriction fragments were analysed with the size and number of DNA fragments.
Statistical Analysis
The Student t-test and Fisher’s-exact test were used to evaluate the differences in the frequency and distribution of Malassezia species between the PV patients and healthy subjects. A p-value ≤ 0.05 was considered statistically significant. A p-value <0.001 was considered as highly significant.
Results
A significantly higher direct KOH results among patients than healthy individuals (94.2% vs 13.3%; p=0.0001) [Table/Fig-1,2]. Also, a significantly higher isolation rate of Malassezia colonies was yielded among PV patients than healthy individuals (71.5% vs 16.7%. p=0.0001) [Table/Fig-2].
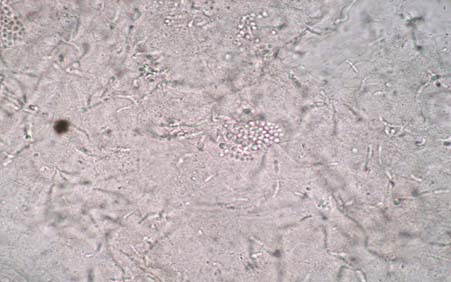
KOH examination of skin scales showing group of round yeast cells and short curved hyphae of Malassezia under x20 magnification.
Demographic and microbiological data of studied PV patients and control individuals.
| Descriptive Data of theStudied Groups | PV Patients(n=137) | HealthyIndividuals(n=30) | p-value |
|---|
| GenderMale (%)Female (%) | 70 (51 %)67 (49%) | 21 (70%)9 (30%) | 0.070** |
| Age (mean±SD) years | 25.5±8.3 | 21.5±9.2 | 0.197* |
| KOH results (no. %) | 129 (94.2%) | 4 (13.3%) | 0.0001** |
| Culture isolates (no. %) | 98 (71.5%) | 5 (16.7%) | 0.0001** |
| Identified Malassezia Species (By both phenotypic and molecular methods) | M. furfur (44)M. globosa (24)M. restricta (10)M. sympodialis (12)M. obtusa (4)M. pachydermatis (4) | M. furfur (3)M. globosa (2) | |
**calculated by Fisher-exact test
*calculated by Student t-test
Mixed cultures were excluded. The positive growth for Malassezia was creamy, moist, pasty colonies on the SDA slants [Table/Fig-3]. Gram stained smears from the colonies were examined for unipolar budding yeast cells of Malassezia.
Sabouraud’s dextrose agar slant covered with olive oil showing shiny and creamy coloured colonies of Malassezia furfur.

By phenotypic methods, five Malassezia isolates from healthy individuals were identified as M. furfur (10%) and M. globosa (6.7%) that were established by PCR [Table/Fig-2].
Out of 137 clinically suspected PV patients, 98 samples were confirmed PV by isolation of Malassezia isolates. Phenotypically, 74 (75.5%) isolates of Malassezia revealed four species as following: M. furfur 38 (51.4%), M. globosa 22 (29.7%), M. restricta 10 (13.5%) and M. pachydermatis 4 (5.4%) while 24 (24.5%) were unidentified isolates. By using PCR technique, all isolates (100%) were identified [Table/Fig-4].
Malassezia species identification by phenotypic methods compared to PCR-RFLP in PV patients.
| Malasseziaspecies | Phenotypic Methods(no. of isolates) | PCR-RFLP(no. of isolates) | p-value |
|---|
| M. furfur | 38 (51.4 %) | 44 (44.9%) | 0.008** |
| M. globosa | 22 (29.7%) | 24 (24.5%) |
| M. restricta | 10 (13.5%) | 10 (10.2%) |
| M. sympodialis | 0 | 12 (12.2%) |
| M. obtusa | 0 | 4 (4.1%) |
| M. pachydermatis | 4 (5.4%) | 4 (4.1%) |
| Total identified | 74 (75.5%) | 98 | 0.0001** |
| Un-identified | 24 (24.5%) | 0 |
| Total | 98 | 98 |
**= calculated by Fishers-exact test
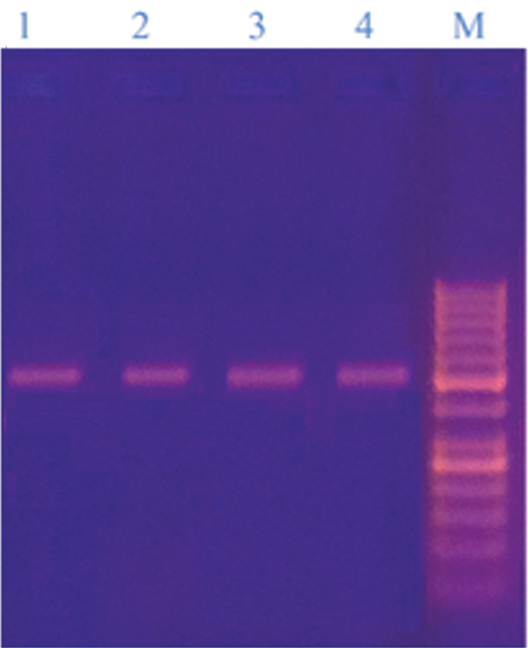
The PCR method produced single PCR product of approximately 580 bp that was further studied by RFLP analysis using Hha1 and BstC1 restriction enzymes [Table/Fig-5].
26S rDNA PCR products of some isolated Malassezia strains (Lanes 1-4: M. furfur, M. sympodialis, M. obtusa, M. pachydermatis (580 bp) and lane M: DNA ladder 50bp, the bands were; 1000, 900, 800, 700, 600, 500, 400, 300, 250, 200, 150, 100, 50. It contains two reference bands (500 and 250 bp);
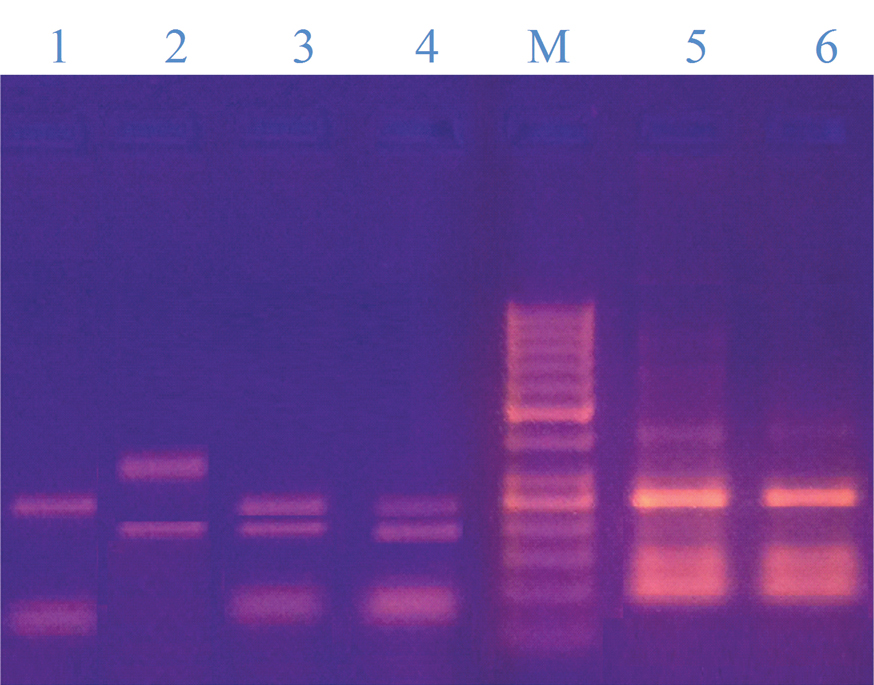
Using Hha1, we could identify five different specie by their specific restriction bands including; M. furfur (bands of 250, (113,109 overlapping), 87 bp), M. globosa (bands of 455, 129 bp), M. restricta (No restriction bands, only band of 580 bp), M. obtusa (bands of 250, 153, 109 bp), M. pachydermatis (bands of 250, 221, 97 bp). On the other hand, colonies of 6 species were identified as either M. sympodialis or M. dermatis (they produced the same digestion pattern, 357, 197 bp) [Table/Fig-6].
Restriction pattern of some isolated Malassezia strains digested by Hha1; Lane 1: M. furfur (250bp, 87bp), lane 2: M. sympodialis (357bp, 197bp), lanes 3&4: M. pachydermatis (250bp, 221bp &97bp), lanes 5&6; M. obtusa (250bp, 153bp& 109bp) and lane M: DNA ladder 50bp, the bands were; 1000, 900, 800, 700, 600, 500, 400, 300, 250, 200, 150, 100, 50. It contains two reference bands (500 and 250 bp);
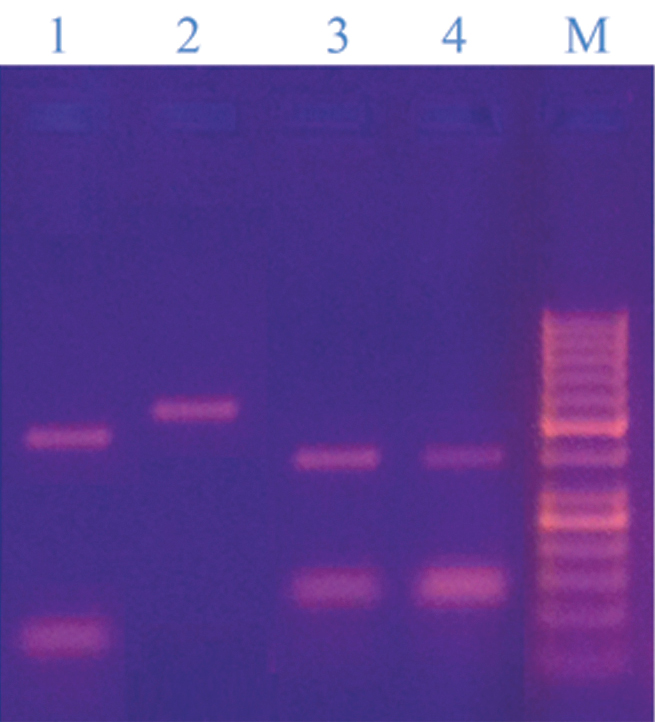
They could be differentiated by using BstC1 enzyme where M. sympodialis produced restriction bands (400, 180bp) and other species were confirmed as following; M. furfur (bands of 400, 180 bp), M. globosa, M. obtusa (No restriction bands, only band of 580 bp), M. restricta, M. pachydermatis (bands of 500, 70 bp) [Table/Fig-7].
Restriction pattern of some isolated Malassezia strains digested by BstC1 enzyme; Lane 1: M. pachydermatis (500bp, 70bp) lane 2: M. obtusa (580bp), lane 3: M. furfur (400bp, 180bp), lane 4: M. sympodialis (400bp, 180bp), and lane M: DNA ladder 50bp, the bands were; 1000, 900, 800, 700, 600, 500, 400, 300, 250, 200, 150, 100, 50. It contains two reference bands (500 and 250 bp).
The tested 98 Malassezia isolates revealed six species: M. furfur 44 (44.9%), M. globosa 24 (24.5%), M. sympodialis 12 (12.2%), M. restricta 10 (10.2 %), M. obtusa and M. pachydermatis 4 (4.1% for each species) as shown in [Table/Fig-4] with significant higher identification rate of isolates by PCR-RFLP compared to phenotypic methods (p=0.008).
The highest frequency of PV (45.9%) was found in patients in the 20-29 age groups with predominant M. furfur, M. globosa and M. obtusa [Table/Fig-8].
Distribution of Malassezia species according to the age groups.
| Malassezia species | Age Groups (year) |
|---|
| < 20 | 20-29 | 30-39 | 40-49 | ≥50 |
|---|
| M. furfur (n=44) | 10 | 21 | 6 | 5 | 2 |
| M. globosa (n=24) | 3 | 13 | 4 | 1 | 3 |
| M. restricta (n=10) | 1 | 3 | 4 | 0 | 2 |
| M. sympodialis (n=12) | 2 | 5 | 0 | 5 | 0 |
| M. obtusa (n=4) | 0 | 2 | 0 | 1 | 1 |
| M. pachydermatis (n=4) | 2 | 1 | 0 | 1 | 0 |
| Total (n=98) | 18(18.4%) | 45(45.9%) | 14(14.3%) | 13(13.3%) | 8(8.1%) |
The higher rate of infection was reported in males (53%) than that in females (47%) which was caused by all species except M. furfur and M. sympodialis which were predominantly seen in females. The most PV patients (64.3%) presented with hypopigmented lesions while only 8.2% patients presented with mixed hypopigmented and hyperpigmented ones. M. furfur, M. globosa, M. sympodialis and M. restricta were predominantly isolated from hypopigmented lesions [Table/Fig-9].
Distribution of Malassezia species according to type of the lesion in both sexes.
| Malassezia species | Type of the Lesion | Gender |
|---|
| Hyperpigmented | Hypopigmented | Mixed | Male | Female |
|---|
| M. furfur (n=44) | 10 | 30 | 4 | 16 | 28 |
| M. globosa (n=24) | 6 | 15 | 3 | 19 | 5 |
| M. restricta (n=10) | 5 | 5 | 0 | 6 | 4 |
| M. sympodialis (n=12) | 4 | 6 | 2 | 5 | 7 |
| M. obtusa (n=4) | 1 | 3 | 0 | 3 | 1 |
| M. pachydermatis (n=4) | 2 | 2 | 0 | 3 | 1 |
| Total (n=98, 100 %) | 27(27.5%) | 63(64.3%) | 8(8.2%) | 52(53%) | 46(47%) |
The most common site affected was the neck and back (25.5%), followed by neck and chest (23.5%), back (20.4%), and then back and chest (13.3%). M. furfur, M. globosa and M. pachydermatis had the most frequency on the neck and back, M. sympodialis on the neck and chest, M. obtusa on back and M. restricta on neck and face [Table/Fig-10]. Regarding healthy control group, all isolates were retrieved from back.
Distribution of Malassezia species in different parts of the lesional skin of PV patients.
| Malassezia species | Site of the Lesion |
|---|
| Neck andBack | NeckandChest | Neck and Face | Neck | Back | Back and Chest | Ch-est |
|---|
| M. furfur (n=44) | 14 | 10 | 2 | 6 | 8 | 3 | 1 |
| M. globosa (n=24) | 7 | 6 | 0 | 2 | 3 | 5 | 1 |
| M. restricta (n=10) | 1 | 0 | 4 | 0 | 3 | 2 | 0 |
| M. sympodialis (n=12) | 1 | 5 | 0 | 0 | 4 | 2 | 0 |
| M. obtusa (n=4) | 0 | 1 | 0 | 1 | 2 | 0 | 0 |
| M. pachyd-ermatis (n=4) | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| Total (n=98, %) | 25(25.5%) | 23(23.5%) | 6(6.1%) | 9(9.2%) | 20(20.4%) | 13(13.3%) | 2(2%) |
Discussion
Pityriasis versicolor is a common recurrent chronic superficial dermatologic infection difficult to eradicate as its pathogenic agents i.e., Malassezia species is endogenous to the skin flora [3,25]. The isolated Malassezia species showed not only a noteworthy difference between healthy individuals and PV patients, but also showed a considerable geographic variation. The Malassezia species distribution among Egyptian population remains unclear.
The conventional methods for diagnosis of PV are direct microscopic examination of skin scrapings treated with 10% KOH and fungal culture. The present study revealed a mycological positivity of 94.2% of skin scrapings of PV patients and 13.3% from healthy control. Using culture, the recovery rate was 71.5% from PV lesions and 16.7% from control individuals. Although, direct microscopic examination of skin scrapings (KOH) is rapid, inexpensive and sensitive for preliminary diagnosis of PV, it lacks a colour contrast and requires a trained eye to interpret. Culture is highly important and specific to differentiate between the Malassezia species phenotypically [26].
As molecular techniques have become common tools to investigate Malassezia species rapidly, accurately and efficiently, we used culture dependent PCR-RFLP to successfully identify Malassezia species in PV patients and healthy individuals [20,27].
Our observations of Malassezia species are concordant with the previous studies that have isolated M. furfur as the predominant species followed by M. globosa and M. sympodialis from PV lesions in Egypt and Nigeria [16,17,28,29], while other studies reported that the most frequently isolated species from PV lesions were M. globosa (53.3% and 47.6%, respectively) followed by M. furfur (25.3% and 41%, respectively) [21,30]. Moreover, Gupta AK et al., found that M. sympodialis was mainly isolated from PV cases in temperate climate and M. globosa was the main species in tropical regions [31]. The distribution of Malassezia species varies with different geographical locations [4]. To our knowledge, this is the first report of the isolation of M. restricta, M. pachydermatis, and M. obtusa from PV in Egypt.
Each Malassezia specie has a specific biochemical and genetic characteristic and specific ecological niche. M. restricta showed a growing frequency as it was isolated from our patients in a higher percentage (10.2%) than others [21,32]. Gaitanis G et al., confirmed that M. restricta is a fastidious organism and is predominantly isolated in non culture based epidemiological studies [8].
In the current study, M. obtusa and M. pachydermatis were isolated from PV patients which is nearly comparable to the results reported by Khosravi AR et al., [33]; whereas, Kaur N et al., [10] isolated them from healthy controls.
It is well known that M. pachydermatis is isolated from patients with close contact with animals like cattle and dogs, live inside or nearby their houses as commonly seen in rural areas [7,10]. Giusiano G et al., explained that M. pachydermatis has been considered only as a transient member of the human cutaneous biota; therefore, its association with PV deserves further investigation [32].
However, Romano C et al., and Rasi A et al., found that M. globosa is the predominant species in temperate climates due to its high lipophilic activity caused by lipases and esterases [19,34]. M. furfur is reported as the main species in Indonesia and Brazil [35,36]. Whereas M. sympodialis and M. globosa were the most frequently isolated species in Canada and Argentina, M. slooffiae and M. restricta were less common in Argentina [31,32].
The differences between the studies may not only be the result of geographical variation in species prevalence, but also due to racial differences, different experimental techniques, hygiene of the subjects, climate, and life styles [9,21].
While El Fangary M and Taha M and Abo Zaid MH et al., found that 7.8% isolates were unidentified and the results of the conventional identification were in agreement with the pattern obtained from 26S rDNA PCR [28,37]. In our study, 24.5% of Malassezia species were not identified phenotypically but they had molecular pattern consistent mainly with M. sympodialis, M. furfur, M. obtusa and to less extent with M. globosa. This could be attributed to the high percentage of these species in our patients and the sensitive PCR-RFLP method we used.
Age group 20-29 years of patients was found to be the most commonly affected with predominant M. furfur, M. globosa and M. obtusa. This is in consensus with Giusiano G et al., who emphasized that the recovery rate of Malassezia in PV is known to be highest in the twenties of both genders when sebaceous glands show greatest activity [32]. Moreover, 21.4 % of PV cases were older than 40 years of age. It may be that subtropical climatic factors of Egypt enable a broader age range of PV [37].
The finding that M. furfur and M. globosa species were highest to be isolated from both healthy skin and PV patients in matched age groups confirms their potential ability to transform from commensal to pathogenic by factors to be further investigated on a large scale.
The effect of sex in predisposition to develop PV is still unclear. A higher rate of infection was observed in males than females with predominance of all species except M. furfur and M. sympodialis, which were predominant in females patients. Giusiano G et al., reported that only M. globosa was more common in female patients [32]. While both isolates of M. furfur and M. globosa were more common in male subjects of control group. Male preponderance seen in the present study may be related to either their higher exposure to factors like high temperature and humidity which are associated with outdoor activities or could be related to increased sebaceous gland activity in men [38].
Similar to other researchers like Ibekwe PU et al., the lesions of 64.3% of studied PV cases corresponded to the hypopigmented type and 8.2% had mixed hypopigmented and hyperpigmented types; whereas, 27.5% had hyperpigmented type [29].
Although Aljebre SH et al., denied any correlation between pigment variation and the type of skin in PV patients [39], Archana BR et al., reported that PV disease has a tendency to be hypopigmented in dark skinned individuals and hyperpigmented in fair skinned individuals [40].
In agreement with Archana BR et al., the present study showed that M. furfur, M. globosa, M. sympodialis and M. restricta were predominantly isolated from hypopigmented lesions [40], whereas M. obtusa and M. pachydermatis were the least.
In this study, we detected a high frequency of patients with multiple lesional sites as the most common site affected was the neck and back, in agreement with Shoeib MA et al., [16]. Most likely the high ambient temperature and humidity of region encourage PV spread [41]. Moreover, areas which remain covered by clothing encourage the development of lesions, supporting the concept that the occlusion of glands plays a role in this disease.
Epidemiologically, we would highlight the frequency of some species at certain body sites. M. furfur, and M. globosa and M. pachydermatis had the most frequency on the neck and back, M. sympodialis on the neck and chest, M. obtusa on back and M. restricta on neck and face. Similar distribution of M. restricta on face was reported by Ibekwe PU et al., study done in Nigeria but different for M. furfur and M. sympodialis which were isolated predominantly from face and back respectively [29]. The distribution of the lesions on various body sites in PV usually parallels the density and activity of sebaceous glands in these areas.
Current study can give an idea about the species present commonly in this locality that may have different sensitivities to antifungal drugs. Further research could facilitate the choosing of appropriate treatment options.
Limitation
We did not use lesional and non lesional samples from same PV patients; hence we could not study the possible effect of individual idiosyncrasy on the results. The relatively low sample size is another limitation. The subjects in this study only represent the population of certain locality in Egypt. A larger study covering all Egyptian Governorates is mandatory for studying the influence of regional factors on the distribution of Malassezia species.
Conclusion
Our results demonstrate that M. restricta, M. pachydermatis, and M. obtusa are pathogenic species for PV in Egyptian patients. The use of PCR-RFLP for identification of Malassezia species helps to deepen the knowledge of regional clinico-epidemiological features of the Malassezia strains that may be useful for selecting sensitive drugs and prevent the occurrence of recurrent or resistant PV infection.
**calculated by Fisher-exact test
*calculated by Student t-test
**= calculated by Fishers-exact test