Infertility in couples is about 15%-20% and can be due to female, male or combined factors. Male factor infertility contributes approximately half of the cases and around 90% or above are characterized by low sperm concentration or by poor quality of spermatozoa in semen [1,2]. Sperms in human are produced by synergy of genes and proteins, through a complicated process called spermatogenesis [3]. Human Y-chromosome is a sex determining chromosome and required for maintenance and development of male germ cells [4,5].
Earlier in 1976, Tiepolo L and Zuffardi O established the strong association between Yq chromosome microdeletions with abnormal spermatogenesis which has given a new insight in the study of Y chromosome long arm for identification and characterization of genetic grounds in spermatogenesis process [6]. After this, Bardoni and colleagues identified the human spermatogenesis locus at Yq 11.23 interval [7]. In 1996, Vogt PH et al., had given evidence that three sub regions (AZFa, AZFb and AZFc) in the AZF associated to specific spermatogenic disruptions [8]. Yq chromosome microdeletions are the second most important genetic cause for spermatogenic arrest in male with infertility after Klinefelter syndrome [9]. AZF region in Yq chromosome contain genes which are involved in spermatogenesis and microdeletions in AZF region leads to spermatogenic defects [10].
Published data showed that there is paucity of AZF microdeletions data in the Gujarati cohort, it is important to have such data as AZF sub regions microdeletions are correlated with spermatogenic failure. Therefore, present study aimed to screen and identifies the Yq chromosome microdeletions in AZF region using PCR as microdeletions cannot be detected by conventional cytogenetic techniques.
Materials and Methods
Study Population
The molecular genetic study was conducted using total 300 patients from Gujarat with oligozoospermia, azoospermia and normozoospermia classified according to the WHO manual 2010 criteria [11], along with two fertile female attending the infertility clinics were incorporated in this study. The study was conducted at Ashok and Rita Patel Institute of Integrated Study and Research in Biotechnology and Allied Sciences (ARIBAS), New Vallabh Vidyanagar, Gujarat, India, during the period of 2014 -2016. Among the 300 patients, 41 were azoospermic, 100 were oligozoospermic and 159 were normozoospermic. The study was approved by the Institutional Ethical Committee of the S.G. Patel Ayurveda Hospital and Maternity Home (Approval No. IEC-3/GJPIASR/2015-16/E/4), Gujarat, India. Written informed consent was signed and collected from all the patients. The study protocols described were in consent as per ICMR human ethical guidelines with respect to human.
The inclusion criteria for our study were oligozoospermia and azoospermia patients aged between 22 to 45 years, couple with infertility having inability to conceive after an unprotected regular intercourse at least for one year, patients with primary infertility attending IVF clinics and male with abnormal seminogram, whereas males with less than 22 years and above 45 years of age, patients with proven immunological, hormonal, chromosomal or other causes of male infertility were excluded from this study.
Morphological Analysis of Spermatozoa
The sperm morphological analysis was carried out by using rapid papanicolau staining kit (Biolab diagnostics, Mumbai). The slides were air dried and observed in light microscope with camera.
DNA Isolation
Semen samples from oligozoospermic and normozoospermic men whereas blood samples from azoospermic patients were collected for AZF microdeletion screening. All the patients instructed through laboratory technicians to follow guidelines for semen sample collection [11] whereas azoospermic blood samples were collected in the EDTA vacutainer tube. Isolation of genomic DNA from the peripheral blood cells and semen samples were carried out using phenol: choloroform method. Qualitative analysis of isolated DNA was performed in 0.8% agarose gel electrophoresis. Quantitative analysis of DNA was carried out by Nanodrop.
Microdeletions Analysis by PCR
Eleven sets of Yq chromosome specific Sequence Tagged Sited (STS) markers were utilized to screen all the three sub-regions of AZF locus using PCR techniques. These STS-primer sets were considered as per European Academy of Andrology, EAA [12]. The following STS markers were used:
AZFa: sY84 and sY86
AZFb: sY121, sY127 and sY134
AZFc: sY153, sY254, sY255, sY1191, sY1197 and sY1291
Internal control: sY14 (SRY)
Independent PCR reactions were designed for all the STS markers for the analysis of microdeletions in AZF region of Yq chromosome. Normal female DNA and reaction mixture with deionized nuclease free water instead of DNA added as negative controls, whereas normal fertile males were used as positive controls. The reaction mixture of PCR was of 25 µL total, which consist of 3 µL genomic DNA templates with concentration of 100 ng and PCR master mix (XcelGen- Premix Taq Version 2.0), nuclease free sterile distilled water, Dimethyl Sulfoxide (DMSO) and STS markers specific primers (Sigma-Aldrich). Amplification of reaction mixture was performed in Eppendorf thermal cycler with initial denaturation at 940C for four minutes followed by 34 cycles of denaturation at 940C for 30 seconds, annealing varying according to STS markers [Table/Fig-1] and extension at 720C for 40 seconds. The final extension was carried out at 720C for three minutes followed by cooling at 40C. PCR products were separated on 2% agarose gel prepared using Tris-acetate-EDTA (TAE) buffer and electrophorized at 100 mA for one hour. Ethidium Bromide (EtBr) stained gel was subjected to UV transilluminator and photographed.
Sequence-tagged sites primers details.
| AZF sub regions | STS markers | Chromosomal Location | Gene Bank Accession no. | Primer sequence (5’-3’) | Annealing temperature (°C) | PCR product size |
|---|
| SRY (Internal Control) | sY14 | Yp11.3 | G38356 | F-GAATATTCCCGCTCTCCGGAR-GCTGGTGCTCCATTCTTGAG | 56 | 472 bp |
| sY84 | Yq11.221 | G12019 | F-AGAAGGGTCTGAAAGCAGGTR-GCCTACTACCTGGAGGCTTC | 57 | 327 bp |
| AZFa | sY86 | Yq11.221 | G49207 | F-GTGACACACAGACTATGCTTCR-ACACACAGAGGGACAACCCT | 57 | 320 bp |
| sY121 | Yq11.222 | G38341 | F-AGTTCACAGAATGGAGCCTGR-CCTGTGACTCCAGTTTGGTC | 61 | 190 bp |
| AZFb | sY127 | Yq11.223 | G11998 | F-GGCTCACAAACGAAAAGAAAR-CTGCAGGCAGTAATAAGGGA | 56 | 274 bp |
| sY134 | Yq11.223 | G12001 | F-GTCTGCCTCACCATAAAACGR-ACCCACTGCCAAAACTTTCAA | 56 | 301 bp |
| sY153 | Yq11.223 | G12004 | F-GCATCCTCATTTTATGTCCAR-CAACCCAAAAGCACTGAGTA | 60 | 139 bp |
| sY254 | Yq11.223 | G38349 | F-GGGTGTTACCAGAAGGCAAAR-GAACCGTATCTACCAAAGCAGC | 60 | 380 bp |
| sY255 | Yq11.223 | G65827.1 | F-GTTACAGGATTCGGCGTGATR-CTCGTCATGTGCAGCCAC | 56 | 124 bp |
| AZFc | sY1191 | Yq11.223 | G73809 | F-CCAGACGTTCTACCCTTTCGR-GAGCCGAGATCCAGTTACCA | 58 | 385 bp |
| sY1197 | Yq11.223 | G67168 | F-TCATTTGTGTCCTTCTCTTGGAR-CTAAGCCAGGAACTTGCCAC | 58 | 453 bp |
| sY1291 | Yq11.223 | G72340 | F-TAAAAGGCAGAACTGCCAGGR-GGGAGAAAAGTTCTGCAACG | 61 | 527 bp |
Results
Morphological Analysis of Spermatozoa
In the present study, 300 semen (azoospemia, oligozoospermia and normozoospermia) samples analyzed for morphological defect. Different types of sperm defects were observed which includes head, mid-piece, tail and mixed defects. In this study, only two oligozoospermia samples (Sample O89 and O91) showed complete (100%) morphological defect.
DNA Isolation
DNA was isolated from all the 259 semen (oligospermic and normozoospermic) and 41 azoospermic blood samples by phenol: choloroform method and subjected to electrophoretic analysis on 0.8% agarose gel. The electrophoretic pattern of DNA isolated, indicates good quality of DNA. DNA quantification of all the semen and blood samples were performed using Nanodrop spectrophotometer.
Azoospermia Patients and Microdeletion
Microdeletion analysis: In the present study, screening of 141 infertile men with the AZF region specific STS markers showed microdeletions in 34 individuals, which accounts for 24.11% of total infertile men analyzed for Yq chromosome microdeletions. Microdeletion pattern of azoospermic individuals and oligozoospermic individuals are represented in [Table/Fig-2,3] respectively. Among the azoospermia patients, 13 (31.41%) out of 41 while 21 (21%) out of 100 oligozoospermia patients showing AZF microdeletions [Table/Fig-4,5,6,7 and 8]. [Table/Fig-9] showing frequencies of microdeletions in each sub-region of AZF. None of the normozoospermic men showed microdeletions in any of the 11 selected STS markers, which were used for the screening for the AZF microdeletions (AZFa, AZFb and AZFc sub-regions).
Results of Y chromosome microdeletions in azoospermia Gujarati males.
| Patients | AZFa | AZFb | AZFc |
|---|
| sY84 | sY86 | sY121 | sY127 | sY134 | sY153 | sY254 | sY255 | sY1191 | sY1197 | sY1291 |
|---|
| A2 | MDel | MDel | - | - | - | - | - | - | - | - | - |
| A3 | - | - | - | MDel | - | - | MDel | - | - | - | - |
| A6 | - | MDel | - | MDel | - | MDel | MDel | - | - | - | - |
| A7 | MDel | MDel | - | - | - | MDel | MDel | - | - | - | - |
| A9 | MDel | - | - | - | - | MDel | - | - | - | - | - |
| A11 | - | - | - | MDel | - | - | - | - | - | - | - |
| A16 | - | - | - | - | - | - | MDel | - | - | - | - |
| A23 | - | - | - | - | - | MDel | - | - | - | - | - |
| A26 | - | - | - | - | - | MDel | - | - | - | - | - |
| A29 | - | - | MDel | - | - | - | - | - | - | - | - |
| A30 | - | - | MDel | - | - | - | - | - | - | - | - |
| A35 | - | - | - | - | - | - | - | MDel | - | - | - |
| A39 | - | - | - | - | - | - | - | - | - | - | MDel |
A: Azoospermia patients and MDel: Microdeletion
Results of Y chromosome microdeletions in oligozoospermia Gujarati males.
| Patients | AZFa | AZFb | AZFc |
|---|
| sY84 | sY86 | sY121 | sY127 | sY134 | sY153 | sY254 | sY255 | sY1191 | sY1197 | sY1291 |
|---|
| O18 | - | - | - | - | - | - | MDel | - | - | - | - |
| O24 | - | - | - | - | MDel | - | - | - | - | - | - |
| O34 | - | - | - | - | - | - | - | - | - | - | MDel |
| O39 | - | - | - | - | - | - | - | - | - | - | MDel |
| O41 | - | - | - | - | MDel | - | - | - | - | - | - |
| O42 | - | - | - | - | - | - | - | - | MDel | - | - |
| O51 | - | - | - | - | - | - | - | - | MDel | - | - |
| O53 | - | - | - | MDel | - | - | - | - | - | - | MDel |
| O56 | - | - | MDel | - | - | - | - | - | - | - | - |
| O57 | MDel | - | - | MDel | - | - | - | MDel | - | - | MDel |
| O62 | - | - | - | - | - | - | - | - | - | MDel | - |
| O67 | MDel | MDel | - | - | - | - | MDel | MDel | - | MDel | - |
| O68 | MDel | MDel | - | - | - | MDel | MDel | MDel | - | - | - |
| O69 | - | - | - | - | - | - | - | - | - | - | MDel |
| O76 | - | - | - | - | - | - | - | MDel | - | - | - |
| O79 | - | - | - | - | - | - | - | - | MDel | MDel | - |
| O83 | - | MDel | - | - | - | - | - | - | - | - | - |
| O89 | - | - | - | - | - | - | - | - | - | - | MDel |
| O90 | - | MDel | MDel | - | - | - | - | - | MDel | MDel | - |
| O91 | - | - | MDel | - | - | - | - | - | - | - | - |
| O92 | - | - | MDel | - | - | - | - | - | - | - | - |
O: Oligozoospermia patients and MDel: Microdeletion
Electrophoretic pattern of microdeletions generated by amplification of DNA using AZFa STS specific primer: a) sY84; b) sY86. N: Normospermic semen samples, O: Oligospermic semen samples, A: Azoospermic blood samples and L: DNA Ladder (100 bp)
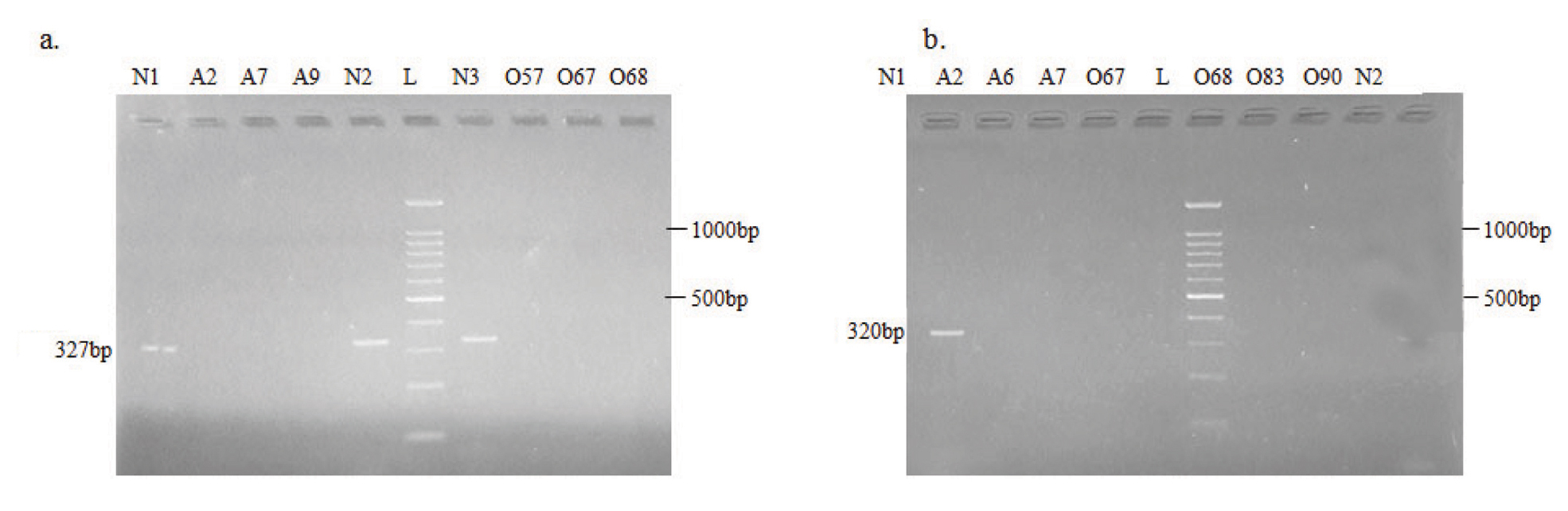
Electrophoretic pattern of microdeletions generated by amplification of DNA using sY121 (Lane 1 to 8), sY127 (Lane 9 to 16) and sY134 (Lane 17 to 19) AZFb STS specific primers.
N: Normospermic semen samples, O: Oligospermic semen samples, A: Azoospermic blood samples and L: DNA Ladder (100 bp)
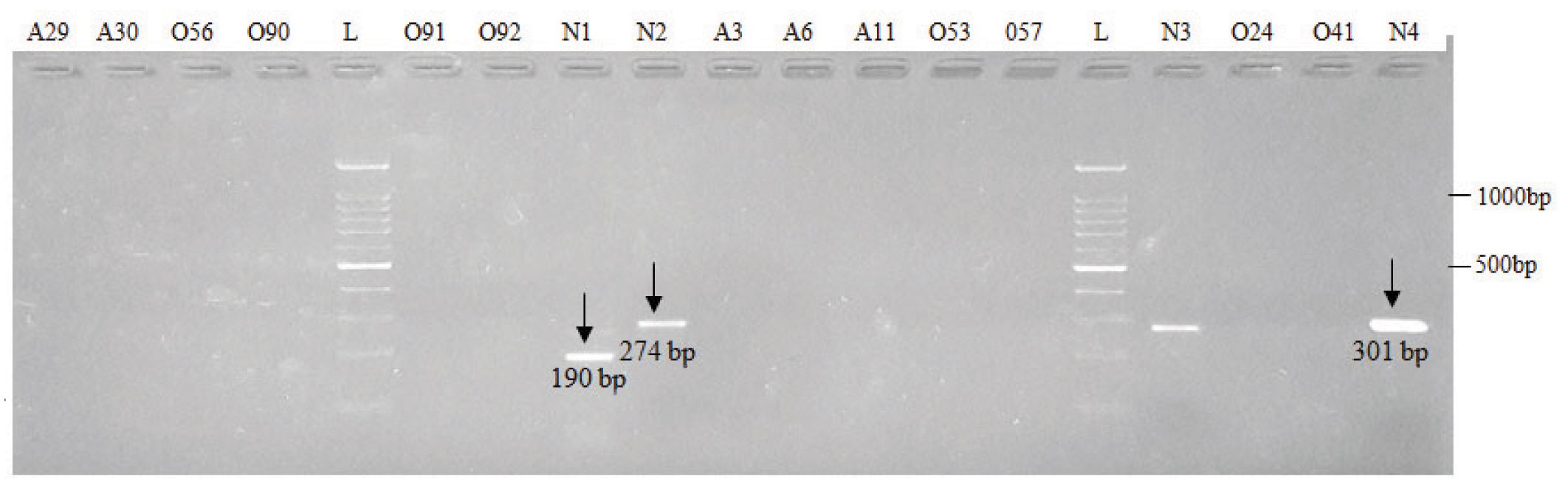
Electrophoretic pattern of microdeletions generated by amplification of DNA using sY153 (Lane 1 to 11) and sY255 (Lane 12 to 20) AZFc STS specific primers.
N: Normospermic semen samples, O: Oligospermic semen samples, A: Azoospermic blood samples and L: DNA Ladder (100 bp)
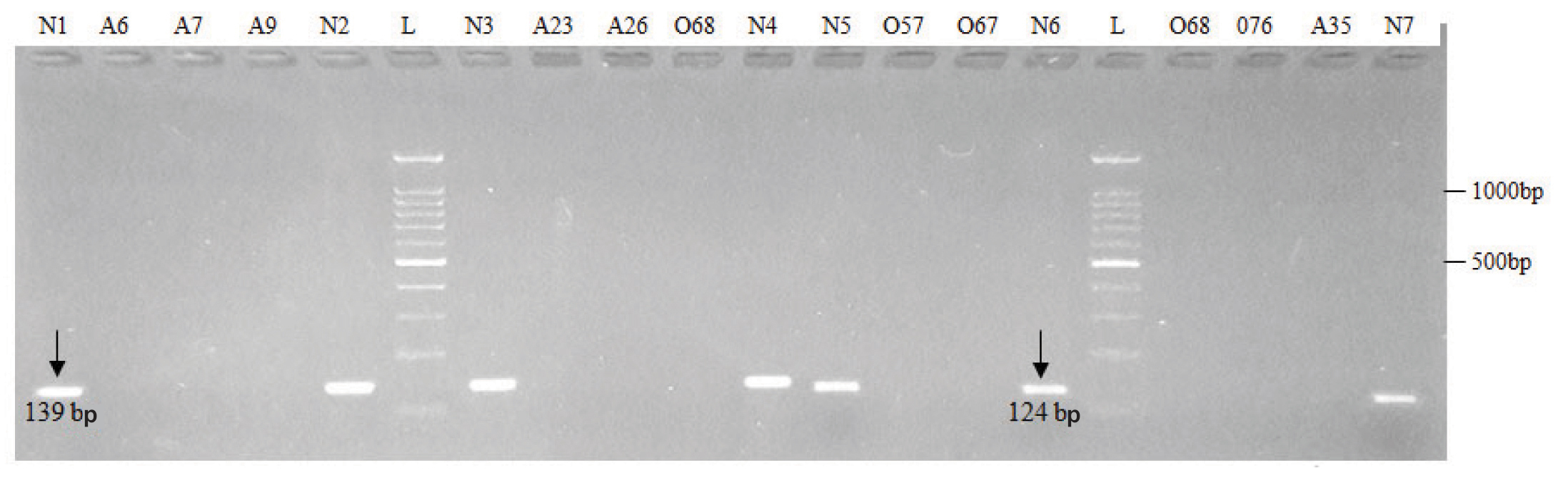
Electrophoretic pattern of microdeletions generated by amplification of DNA using sY254.
(Lane 1 to 12) and sY1191 (Lane 13 to 20) AZFc STS specific primers.
N: Normospermic semen samples, O: Oligospermic semen samples, A: Azoospermic blood samples and L: DNA Ladder (100 bp)
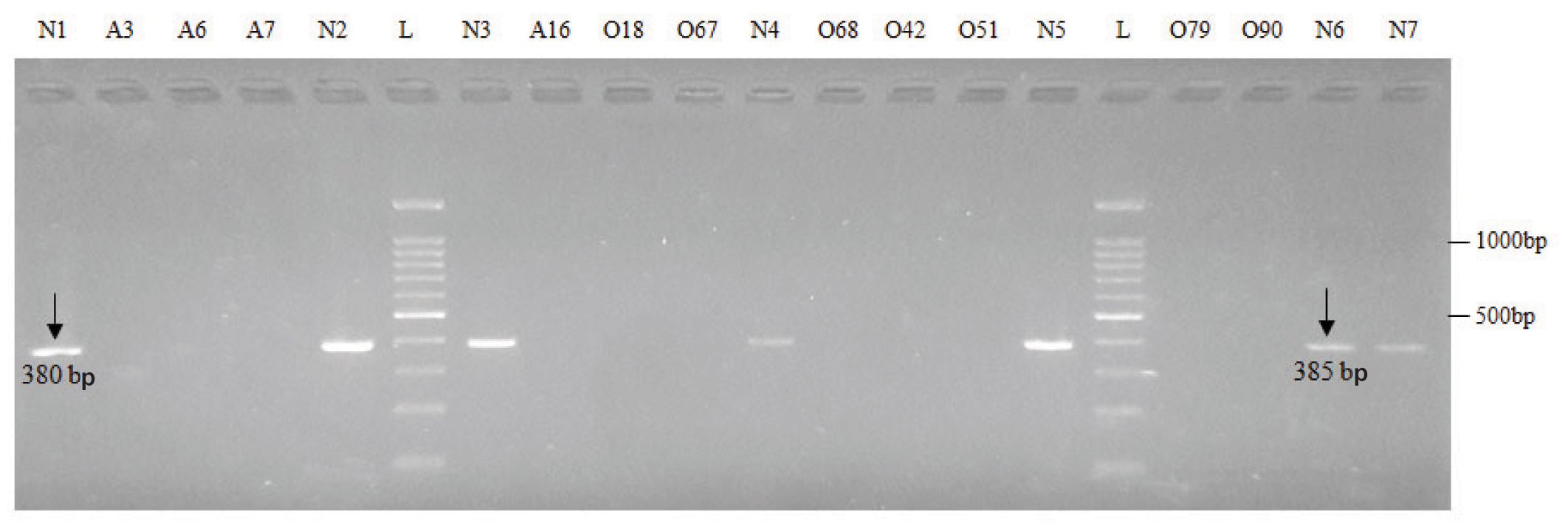
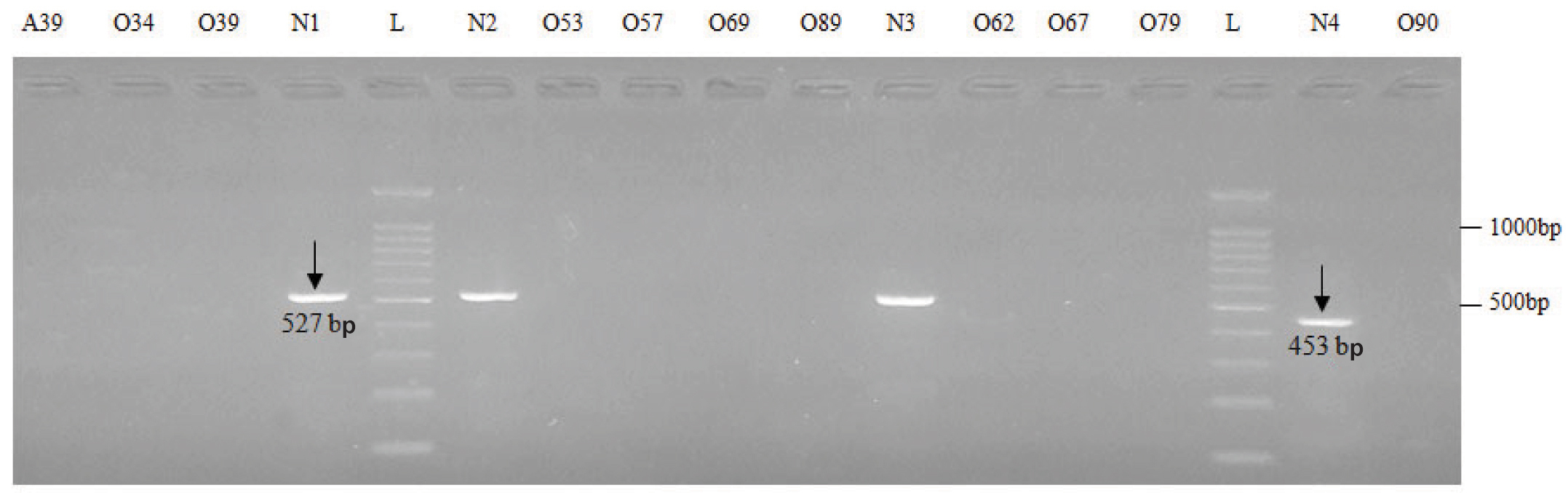
Electrophoretic pattern of microdeletions generated by amplification of DNA using sY1291 (Lane 1 to 10) and sY1197 (Lane 11 to 17) AZFc STS specific primers.
N: Normospermic semen samples, O: Oligospermic semen samples, A: Azoospermic blood samples and L: DNA Ladder (100 bp)
Frequency distribution of Y chromosome microdeletion subtypes in infertile males.
| Class | AZFa | AZFb | AZFc |
|---|
| sY84 | sY86 | sY121 | sY127 | sY134 | sY153 | sY254 | sY255 | sY1191 | sY1197 | sY1291 |
|---|
| Azoospermia (n=41) | 3 (7.3%) | 3 (7.3%) | 2 (4.9%) | 3 (7.3%) | 0 (0%) | 5 (12.2%) | 4 (9.8%) | 1 (2.4%) | 0 (0%) | 0 (0%) | 1 (2.4%) |
| Oligozoospermia (n=100) | 3 (3%) | 4 (4%) | 4 (4%) | 2 (2%) | 2 (2%) | 1 (1%) | 3 (3%) | 4 (4%) | 4 (4%) | 4 (4%) | 6 (6%) |
| Normozoospermia (n=159) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
Discussion
After Klinefelter syndrome, the AZF microdeletion in Yq chromosome is considered as second cause of spermatogenic arrest [12]. There was variable but acceptable percentage of sperm morphological defects (head, mid-piece, tail and mixed defects) as per the WHO, 2010 guidelines (upto 96% acceptable). The morphological defects observed in all the sperms in sample number O89 and O91. Both the samples are of oligozoosoermia and there is a deletion of only single STS sY1291 in O89 and sY121 in O91. Interestingly, sample O67 and O68 had microdeletion in five STS out of 11 but they had acceptable sperm morphological defects. This proves that there were no association between sperm morphological defect and Yq microdeletion in the present study. In our study, we determined the prevalence of AZF microdeletions in Yq chromosome for infertile men, with the aim to find out the correlation between AZF microdeletions with male infertility in the population of Gujarat. A bunch of genes on the Yq chromosome regulates the spermatogenesis. In recent years, Yq chromosome microdeletions have appeared as a major cause of male factor infertility. Yq chromosome microdeletion is reported as second most frequent genetic cause in male factor infertility after Klinefelter syndrome [13].
It was also reported that microdeletions in AZF region can lead to a variable phenotype with a significant reduction in sperm count and increased loss of germ cells and progressive decline in semen quality. According to EAA guidelines for diagnostic testing [12,14] and most recent works [15,16], we have choose STS markers for our study.
PCR is a very sensitive technique to detect constitutional Yq microdeletions. A total of 259 semen samples and 41 blood samples of fertile and infertile men were screened for sub-microscopic Yq chromosome deletions. Molecular analysis of AZF (AZFa, AZFb, AZFc) sub regions was carried out using eleven sY84, sY86, sY121, sY127, sY134, sY153, sY254, sY255, sY1191, sY1197 and sY1291 STS specific primers and additionally one internal marker sY14 (SRY). University of California, Santa Cruz (UCSC) genome browser and male specific region of the Human Y chromosome (MSY) breakpoint mapper is used to check the PCR product size of particular STS markers. In the present study, thirty four (24.11%) out of 141 infertile men showed microdeletions in Yq chromosome of AZFa, AZFb and AZFc sub-regions. No microdeletions were identified in any of the control males. The frequency of Yq microdeletion in India has shown vast variation ranging from 0 to 36%. For such vast variations in Yq microdeletion frequency is due to many reasons like geographical as well as racial variations [17], fluctuation in sample size [18] and number and types of STS markers used [19-21]. In Indian cohort, the very first report on Yq microdeletion was studied by Babu SR et al., which reported frequency of 5% [22]. In present study, the Yq microdeletion frequency is higher (24.11%) in Gujarati population which is very similar to 24.7% that was recorded by Abilash VG et al., 2010 in Southern East Indian population [23]. Yq microdeletion frequency recorded by Suganthi R et al., in Southern Indian population was 1.5 times more than this study [24]. Mitra A et al., found slightly higher frequency of Yq microdeletion (28.6%) in there study on Indian population [25].
The frequency of microdeletion reported in our study matches with other published reports also. For example Hofherr SE et al., reported frequency of 24% and Omrani MD et al., reported a frequency of 24.2% in infertile male patients [26,27]. Yq chromosome microdeletions results in this study are higher than the studies reported by some other authors. For example, Behulova R et al., reported 14.07% of AZF microdeletion in Northeast China and Fu L et al., reported 10.80% of AZF microdeletion in Chinese infertile men [28,29]. However, the frequency of AZF microdeletions reported in our study is lower than the frequency reported by Arruda JT et al., Elhawary NA et al., and Elsaid H et al., which is 43.48%, 39.28% and 37.5% respectively [30-32]. The frequencies of all the three regions are shown in [Table/Fig-10]. Two azoospermia samples A2 and A7 showed deletions in both AZFa specific markers. Two oligozoospermia patients (O67 and O68) showed microdeletions in five STS markers out of eleven with respect to AZFa and AZFc. In the present study population, it has been observed that the AZFc region deletion frequency is comparatively higher followed by AZFb whereas lowest frequency is found in AZFa region. Among the 41 azoospemia patients, 4 (9.75%) patients in AZFa, 5 (12.19%) patients in AZFb and 9 (21.95%) patients in AZFc carry microdeletions. Similarly, out of 100 oligozoospermia 5 (5%) patients in AZFa, 8 (8%) patients in AZFb and 15 (15%) patients in AZFc showed microdeletions. In azoospermic patients, two (A6 and A7) showed deletion of four STS-markers, three patients (A2, A3 and A9) showed deletion of two STS-markers and rest eight patients showed deletion only one STS-marker. In oligozoospermic patients, two (O67 and O68) showed deletion of five STS-markers, two patients (O57 and O90) showed deletion of four STS-markers, two (O53 and O79) showed deletion of two STS-markers and remaining 15 showed deletion of only one STS-markers [Table/Fig-2,3].
Number of infertile and fertile subjects within different sub-regions of AZF.
| Group | AZFa | AZFb | AZFc |
|---|
| Infertile group (n=141) | 9/141 (6.38%) | 13/141 (9.22%) | 24/141 (17.02%) |
| Fertile group (n=159) | 0 (0%) | 0 (0%) | 0 (0%) |
When compared overall results with the seminogram, the present study results and the evidence found in various studies suggest that the infertile men sperm concentration less than 15x106/ml [11], should be screened for AZF microdeletions, which help clinician to avoid expensive ART treatments. Comparing overall result together, it could be found that microdeletions in Yq chromosome AZFa, AZFb and AZFc sub-region in the infertile male is one of the main causative factor of infertility. The microdeletion in the azoospermia factor region (AZF) of Y-chromosome was scanned in the present study using PCR. It is very powerful technique where sensitivity and specificity depends on purity of DNA. Therefore, enough care should be taken during the collection and transport of the semen and blood samples as well as during isolation of DNA to avoid possibility of contamination. Additionally, microdeletion pattern reproduced atleast thrice by the laboratory along with Y-specific internal STS markers (sY14). In future, more number of STS markers in different sub-regions of AZF and large number of infertile patients are required for better understanding of genetic background and pathogenic significance in infertile man.
Conclusion
The frequency of microdeletion of the AZFc sub region was found to be higher followed by AZFb and AZFa sub region and the prevalence of AZF deletion in this Gujarati study population, was 24.11%. The Yq chromosome microdeletion analysis is highly recommended for oligozoospermia and azoospermia infertile males of Gujarat to explore appropriate assisted reproductive techniques. It also lowers down the risk of transmission of AZF microdeletion to future generations. Frequency varies among different reports, this may be due to selection criteria of populations, different diagnostic methods and/or selection of STS markers by researchers.