First Detection and Characterization of Streptococcus dentapri from Caries Active Subject
Hamzah Abdulrahman Salman1, R. Senthil Kumar2, N. Chaitanya Babu3, Khalid Imran4
1 Research Scholar, Department of Microbiology, J.J. College of Arts and Science, Affiliated to Bharathidasan University, Pudukkottai, Tamil Nadu, India.
2 Associate Professor, Department of Microbiology, J.J. College of Arts and Science, Affiliated to Bharathidasan University, Pudukkottai, Tamil Nadu, India.
3 Professor, Department of Oral Pathology, The Oxford Dental College, Affiliated to Rajiv Gandhi University, Bengaluru, Karnataka, India.
4 Research Associate, Department of Life Sciences, Nucleobase Life Sciences Research Laboratory and Associate Professor, Department of Biotechnology, Krupanidhi Degree College Bengaluru, Karnataka, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Khalid Imran, Research Associate, Department of Life Sciences, Nucleobase Life Sciences Research Laboratory, No: 1056, I Stage, IV Block, V Cross, HBR Layout, Hennur Bellary Road, Bengaluru-560043, Karnataka, India.
E-mail: qhalidimran@gmail.com
Introduction
Mutans streptococci (MS) are a group of oral bacteria generally regarded as the principal agents in the pathogenesis of dental caries.
Aim
The study aimed was characterize S. dentapri based on phylogenetic analysis and phenotypic methods from Caries Active Subject.
Materials and Methods
While sequencing MS species which were isolated from 65 caries active subjects, one strain of S. dentapri was detected. Dental plaque samples were processed and cultured on mitis salivarius bacitracin agar. S. dentapri was characterized using phylogenetic analysis, colony morphology characterization and biotyping.
Results
Among the study population, one strain designated as H14 was identified as S. dentapri by 16S rDNA sequencing. Morphologically, S. dentapri could not differentiate from other species of MS. S. dentapri H14 demonstrated biotype II biochemical characteristics of MS. The phylogenetic analysis showed S. dentapri is closely related to S. macacae.
Conclusion
The study concludes that S. dentapri can inhabit the human oral cavity and therefore further investigations are warranted to determine its role in caries.
Biotyping, Colony morphology, Mutans streptococci, 16S rDNA sequencing
Introduction
Dental caries is one of the oldest, multifactorial and transmissible infectious disease, affecting all the age groups of humans [1,2]. Dental caries is a result of an imbalance of the metabolic activity in the dental plaque [3]. Despite the complex diversity of oral flora, Mutans streptococci (MS) have been widely considered as the major cariogenic bacteria [4]. MS have been classified into 14 species, namely Streptococcus mutans, S. sobrinus, S. downei, S. dentapri, S. criceti, S. dentirousetti, S. devriesei, S. ferus, S. macacae, S. orisratti, S. orisuis, S. ratti, S. troglodytae and S. ursoris [5]. However, three more novel species have been recently added to MS group, i.e., S. dentasini, S. oriloxodontae and S. orisasini [6,7]. Among MS species, S. mutans and S. sobrinus are the most isolated species from human dental caries [4,8].
Recently, Takada K et al., reported a novel species of MS, S. dentapri, isolated from the oral cavity of wild boar [9]. Since then there are no reports of detection of S. dentapri, though extensive research has been done on MS. However, while studying clinical strains of MS isolated from caries active subjects, one strain of S. dentapri was detected. In this background, the objective of the study was to characterize S. dentapri based on phylogenetic analysis and phenotypic methods.
Materials and Methods
The present observational study was conducted at Nucleobase Life Sciences Research Laboratory, Bengaluru, India, from March 2013 to March 2016. This study was approved by the Ethical Committee of the PMNM Dental College, Bagalkot, affiliated to Rajiv Gandhi University of Health Sciences, Karnataka, India. The plaque samples from 65 caries active subjects aged between 35 to 44 years were collected using sterile wooden toothpicks and then transferred to vials containing 1 ml sterile phosphate buffer saline (HiMedia, India). The vials were vortexed for one minute and diluted by 100-fold in 1X phosphate buffer saline. The processed plaque samples were then plated on Mitis Salivarius Bacitracin (MSB) agar (HiMedia, India) and anaerobically incubated at 37°C for 48 hours [10,11]. Obtained cutures were subjected to sequencing.
Genomic DNA isolation: The bacterial DNA was isolated according to the methodology described by Moreira M et al., with a few modifications [12]. Briefly, 1 ml of 24-hours-old culture broth was centrifuged at 49,000 g for two minutes and the supernatant was discarded. The pellet was washed with sterile distilled water and then centrifuged at 49,000 g for two minutes. Approximately 675 μl of TE buffer (100 mM Tris-Cl and 100 mM Ethylenediaminetetraacetic acid), 1.4 M NaCl, 1% Cetyl Trimethyl Ammonium Bromide (CTAB) and 0.03 μg/μl proteinase K (Sigma, India) was added and incubated at 37°C for 30 minutes. Almost 75 μl of sodium dodecyl sulfate (20%) was added and incubated at 65°C for two hours. After incubation, the solution was centrifuged at 49,000 g for two minutes and the supernatant was collected in a sterile Eppendorf tube. Chloroform: isoamyl alcohol (24:1 v/v) was added and centrifuged at 49,000 g for seven minutes. The aqueous phase was transferred to another vial and the solution was homogenized by adding isopropyl alcohol (0.6 v/v) and incubated at room temperature for one hour. After centrifuging at 49,000 g for seven minutes, 70% of cold ethanol was added. The solution was centrifuged at 49,000 g for seven minutes and the supernatant was discarded. The vial was left open in air chamber to dry the pellet and later suspended in 25 μl of ultra-pure water and stored at -20°C until further use. The concentration of eluted bacterial DNA was determined by measuring at A260 using UV spectrophotometer (Sartorius stedim biotech, Germany) and later the purity of DNA was assessed from A260/A280 ratio.
16S rDNA sequencing: The clinical isolate was identified by 16S rDNA sequencing, the PCR was performed using universal primer 16S FP5’-AGAGTTTGATCCTGGCTCAG-3’, 16S RP5’-AAGGAGGTGATCCAGCCGCA-3’ [11]. The following conditions were maintained: initial denaturation 94°C for two minutes, denaturation at 94°C for 50 seconds, annealing at 48°C for 30 seconds, extension at 72°C for 90 seconds and final extension at 72°C for six minutes. The GenBank accession number was obtained from the National Centre for Biotechnology Information (NCBI) database after the submission of the 16S rDNA sequences.
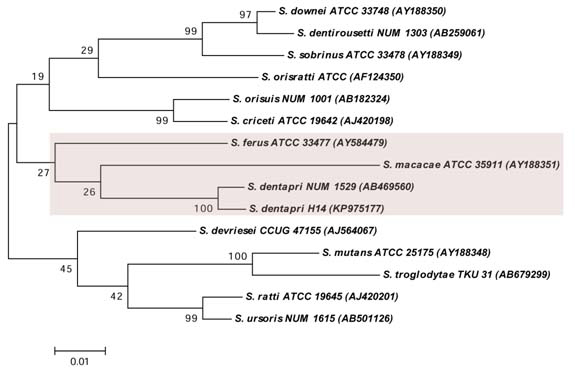
Phylogenetic analysis: The phylogenetic analysis of the S. dentapri H14 was performed along with 14 members of MS species, whose determined 16S rDNA sequences were retrieved from GenBank nucleotide sequence database. The phylogenetic tree was performed based on the Kimura 2-parameter model [13]. The tree was constructed by maximum likelihood method. The tree with the highest log based likelihood (-5486.8001) was considered for the analysis. The percentage of the tree in which the related taxa grouped together was depended on 1000 resampling. Primary tree in the heuristic search was achieved by applying the neighbour-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood approach and the analyses were carried out in MEGA 6 [14]. Based on the measurement of branch lengths in the number of replacements per site, the tree was drawn to scale. The acquired positions with less than 85% site coverage were removed, e.g. fewer than 15% absent data, alignment gaps and vague bases. There were a total of 1423 positions in the final data set.
Biochemical characterization: Biochemical characterization was performed as described by Takada K et al., with a few modifications [9]. The biotype of S. dentapri was determined as described by Yoo SY et al., [10]. A phenol red broth base (HiMedia, India) was used as the basal medium for the fermentation of mannitol, melibiose, salicin, trehalose, sorbitol, inulin, raffinose and lactose discs (HiMedia, India). Arginine dihydrolase and esculin fermentation broth (HiMedia, India) were also employed in biochemical characterization. After incubation at 37°C for 48 hours the tubes were observed for colour change to distinguish the positive and negative reaction. Reproducibility and reliability of the results were affirmed by repeating the experiments in triplicate.
Results
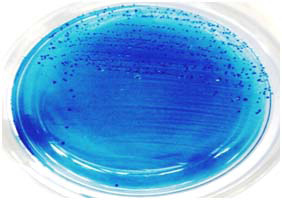
Among the 65 clinical isolates, one isolate designated as H14 was detected as S. dentapri by 16S rDNA sequencing. The GenBank accession number provided by NCBI for the strain H14 was KP975177. The 16S rDNA sequences of the H14 strain demonstrated highest similarity with S. dentapri NUM 1529 (99 %). From the phylogenetic analysis, it was observed that S. dentapri is closely related to S. macacae than other members of MS [Table/Fig-1]. The colony characteristics of S. dentapri H14 was found to be small, raised, irregular, rough, dark-blue in colour and glistening on the MSB agar [Table/Fig-2]. The S. dentapri H14 expressed biotype II biochemical characterization of MS. The biochemical characteristics of S. dentapri are outlined in [Table/Fig-3] and the biochemical test results are represented in [Table/Fig-4].
Phylogenetic analysis depicting the relationship of S. dentapri with other MS species based on the 16S rDNA sequences. The tree constructed by maximum likelihood method generated using Kimura-2-parameter model in MEGA 6.0 software. The accession numbers are shown in parenthesis. The numbers above the branches referred to bootstrap values based on 1000 resampling.
Colonies of S. dentapri H14 on MSB agar.
Biochemical characteristics of S. dentapri, S. macacae and S. mutans.
| SL. No. | Isolate | Mannitol | Melibiose | Salicin | Trehalose | Sorbitol | Inulin | Raffinose | Lactose | Arginine | Esculin |
|---|
| 1. | S. dentapri H14 | + | + | + | + | + | - | + | + | + | + |
| 2. | S. dentapri NUM 1529 | + | - | + | + | + | ND | - | - | - | + |
| 3. | S. macacae | + | - | ND | + | + | - | + | + | - | - |
| 4. | S. mutans | + | +/- | + | + | + | + | + | + | - | + |
1, data obtained in this study; 2, data obtained from [9]; 3 and 4, data obtained from [19]; +, fermentation positive; -, fermentation negative; ND, Not determined.
Biochemical tests of S. dentapri H14.
Mn: Mannitol, Mb: Melibiose, Sa: Salicin, Tr: Trehalose, Sb: Sorbitol, In: Inulin, Rf: Raffinose, La: Lactose, Ar: Arginine, Es: Esculin Fermentation Broth.

Discussion
To the best of our knowledge, this is the first reported study of isolation of S. dentapri from human active caries. In context to the morphological characterization, S. dentapri could not be differentiated from other MS species. In contrast, molecular identification based on 16S rDNA sequencing showed a high level of sensitivity for detection of MS species [15]. Phylogenetically, S. dentapri is closely related to S. macacae when compared with other MS [Table/Fig-1] which is in agreement with the previous study [9].
In the present study, the clinical strain of S. dentapri displayed different biochemical characteristics when compared with the type strain S. dentapri NUM 1529 [Table/Fig-3]. The reason for the strain metabolic variability might be due to heterogeneity of S. dentapri or dietary habits of the host. S. dentapri H14 demonstrated biotype II characteristics which was earlier detected in few strains of MS species namely S. ratti, S. downei and S. mutans [10,16,17]. However, within a species, strains can be differing with respect to biochemical characteristics and the same strains can exhibit unreliable metabolic activity [18]. The biochemical characteristics of S. dentapri were also compared with the results of an earlier study with reference to S. macacae and S. mutans [Table/Fig-3] [9,19]. S. dentapri and S. macacae displayed contrary biochemical results for melibiose fermentation and arginine hydrolysis, while S. dentapri and S. mutans demonstrated variation for inulin fermentation and arginine hydrolysis.
Conclusion
As this is the first finding of S. dentapri in caries active subject, this study recommends further investigations to be carried out for determining the virulence factors of S. dentapri.
1, data obtained in this study; 2, data obtained from [9]; 3 and 4, data obtained from [19]; +, fermentation positive; -, fermentation negative; ND, Not determined.
[1]. Ferreira Zandoná A, Santiago E, Eckert GJ, Katz BP, de Oliveira SP, Capin OR, The natural history of dental caries lesions: a 4- year observational studyJ Dent Res 2012 91:841-46. [Google Scholar]
[2]. Ravishankar PL, Jayapalan CS, Gondhalekar RV, Krishna BJ, Shaloob KM, Ummer PF, Prevalence of dental caries and oral hygiene status among school going children: an epidemiological studyJ Contemp Dent Pract 2013 14:743-46. [Google Scholar]
[3]. Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D, Dental caries from a molecular microbiological perspectiveCaries Res 2013 47:89-102. [Google Scholar]
[4]. Loesche WJ, Role of Streptococcus mutans in human dental decayMicrobiol Rev 1986 50:353-80. [Google Scholar]
[5]. Toit MD, Huch M, Cho GS, Franz CM, The genus StreptococcusIn: Lactic Acid Bacteria: Biodiversity and Taxonomy. Holzapfel WH, Wood BJB, editors 2014 John Wiley & Sons:457-505. [Google Scholar]
[6]. Takada K, Saito M, Tsudukibashi O, Hiroi T, Hirasawa M, Streptococcus orisasini sp. nov. and Streptococcus dentasini sp. nov., isolated from the oral cavity of donkeysInt J Syst Evol Microbiol 2013 63:2782-86. [Google Scholar]
[7]. Shinozaki-Kuwahara N, Saito M, Hirasawa M, Takada K, Streptococcus oriloxodontae sp. nov., isolated from the oral cavities of elephantsInt J Syst Evol Microbiol 2014 64:3755-59. [Google Scholar]
[8]. Oda Y, Hayashi F, Okada M, Longitudinal study of dental caries incidence associated with Streptococcus mutans and Streptococcus sobrinus in patients with intellectual disabilitiesBMC Oral Health 2015 15:102 [Google Scholar]
[9]. Takada K, Hayashi K, Sata Y, Hirasawa M, Streptococcus dentapri sp. nov., isolated from the wild boar oral cavityInt J Syst Evol Microbiol 2010 60:820-23. [Google Scholar]
[10]. Yoo SY, Park SJ, Jeong DK, Kim KW, Lim SH, Lee SH, Isolation and characterization of the mutans streptococci from the dental plaques in KoreansJ Microbiol 2007 45:246-55. [Google Scholar]
[11]. Salman HA, Senthilkumar R, Antibacterial activity of Annona squamosa L. and Annona reticulata L. against clinical isolates of mutans streptococci the causative agents of dental cariesAsian J Pharm Clin Res 2015 8:152-55. [Google Scholar]
[12]. Moreira M, Noschang J, Neiva IF, Carvalho Y, Vicente VA, Methodological variations in the isolation of genomic DNA from Streptococcus bacteriaBraz Arch Biol Technol 2010 53:845-49. [Google Scholar]
[13]. Kimura M, A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequencesJ Mol Evol 1980 16:111-20. [Google Scholar]
[14]. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, MEGA6: Molecular Evolutionary Genetics Analysis version 6.0Mol Biol Evol 2013 30:2725-29. [Google Scholar]
[15]. Salman HA, Senthilkumar R, Identification and antibiogram profile of Streptococcus mutans and Streptococcus sobrinus from dental caries subjectsJ App Pharm Sci 2015 5:54-57. [Google Scholar]
[16]. Coykendall AL, Alan L, Classification and identification of the viridans streptococciClin Microbiol Rev 1989 2:315-28. [Google Scholar]
[17]. Yoo SY, Kim KJ, Lim SH, Kim KW, Hwang HK, Min BM, First isolation of Streptococcus downei from human dental plaquesFEMS Microbiol Lett 2005 249:323-36. [Google Scholar]
[18]. Lal D, Verma M, Lal R, Exploring internal features of 16S rRNA gene for identification of clinically relevant species of the genus StreptococcusAnn Clin Microbiol Antimicrob 2011 10:1-28. [Google Scholar]
[19]. Whiley RA, Beighton D, Current classification of the oral streptococciOral Microbiol Immunol 1998 13:195-216. [Google Scholar]