Introduction
Helicobacter pylori as a human common gastroduodenal pathogen colonizes the gastric mucosa more than half of the world’s population and induces disorders ranging from gastritis and Peptic Ulcer Disease (PUD) to gastric malignancies [1,2]. Many genes are involved in virulence of H. pylori, which includes cell wall synthesis gene glmM (ureC) one of the key genes for molecular confirmation of all H. pylori strains and is different from ureA (for urease production) gene [3,4].
Adherence of H. pylori to the gastric epithelium cells is believed to be a major process for stomach inflammation. About 32 H. pylori Outer Membrane Proteins (OMPs) as adhesins are involved in this process [5]. These adhesins as virulence factors have been recognized in the Hop (Helicobacter outer membrane protein) group [6,7]. Three of the most common Hop adhesins are blood group antigen-binding adhesin (BabA or HopS), outer inflammatory protein A (OipA or HopH) and sialic acid-binding adhesin (SabA or HopP). ABO blood group and Lewis B (leb) antigens on erythrocytes and gastric mucosa are receptors for babA [8,9]. The babA gene contains babA1 and babA2 alleles; but, due to an insertion at the 3’ end of the gene [10], only the babA2 product is functional for binding [9,11]. It has been shown that 70% of Western H. pylori isolates associated with severe diseases were harbored babA2 [11]. BabA2- mediated strong adhesion to gastric epithelial cells might enhance delivery of CagA (Cytotoxin-Associated Gene A) and VacA (Vacuolating Cytotoxin A) cytotoxins into the host cells; therefore, may be associated with peptic ulcer and gastric cancer [12].
Apart from the role of bacterial colonization, OipA is a powerful inducer of Interleukin-8 (IL-8) secretion by gastric epithelia, which results in elevated levels of gastric inflammation. This process is due to phosphorylation of multiple signaling pathways that interact to cag PAI (the cag pathogenicity island)-related pathways [13]. To date, the specific receptor for OipA has been remained unknown [6,9,14].
H. pylori infection induces expression of sialylated glycans, namely sialyl-Lewis x/a antigens (sLeX and sLea) in the gastric mucosa. The expression of these antigens, in particular sLea, has been reported to be associated with gastric malignancies in both developed and developing countries. Hence, further investigations on SabA in developing countries has been suggested [15,16]. Both oipA and SabA genes, together with babA in some strains [16], are regulated by phase variation (switch “off”=non functional and switch “on”= functional), through a slipped-strand mispairing mechanism based on the number of calcitonin or Cysteine-Threonine (CT) dinucleotide repeats in the 5’ coding region [7,17-19].
The association of H. pylori different genotypes with clinical manifestations varies within societies. However, investigation on putative virulence factors is crucial to provide useful clinical data. Therefore, as there is no data available about geographic prevalence of H. pylorioipA, SabA and babA2 genotypes from our region, we decided to assess the frequency of these genes among H. pylori clinical isolates from Iran.
Materials and Methods
Study Population
In this cross-sectional study 100 H. pylori isolates were obtained from antral biopsies of 318 patients referred for endoscopy in Shiraz Faghihi Hospital, Southwest of Iran, from January to May 2014 [20]. The H. pylori positive subjects were classified on the basis of endoscopic and histopathological findings into gastritis (Ga, n= 63), Gastric Ulcer (GU, n=15), Duodenal Ulcer (DU, n= 13) and Non-Ulcer Dyspepsia (NUD, n=9) groups. According to written informed consent, no patient had received anti-H. pylori therapy and endoscopy within the last month. The study was approved by the Ethics Committee of Shiraz University of Medical Sciences (EC-9379-7059).
H. pylori Identification and PCR-based Genotyping
Antral biopsy samples were processed and cultured as previously described [20]. Briefly, transported specimens within Brucella Broth (Quelab, Montreal, QC, Canada) to the Microbiology laboratory, were plated onto Columbia agar (Merck kGaA, Darmstadt, Germany) supplemented with 7% sheep blood and specific antibiotics (vancomycin 10 mg, polymyxin B 2500 IU, trimethoprim 5 mg and amphotericin B 2.5 mg/L). After 5-10 days, incubation at 37°C under microaerobic conditions (5% O2, 10% CO2 and 85% N2), identification of H. pylori was done based on typical morphology in Gram staining and conventional biochemical tests including catalase, oxidase and urease production tests. Indeed, DNA extraction from all 100 H. pylori pure cultures and storage conditions were carried out as previously described [20].
PCR was used for detection of H. pylori-specific glmM (for molecular confirmation) and presence of oipA, SabA and babA2 genes. Primer sets used were provided from the published literature [1,4,21,22]. Each PCR reaction was done in a final volume of 25 μl containing 1X PCR buffer, 400 nM of each primer, 1.5 mM MgCl2, 200 μM of dNTPs mix, 1U Taq DNA polymerase and 200 ng (2 μl) of genomic DNA. PCR amplifications were performed in a thermocycler (Eppendorf AG 22331; Eppendorf, Hamburg, Germany) under the specific conditions [Table/Fig-1]. The amplicons were resolved on 1.5% agarose gel with ethidium bromide and visualized under U.V light. In all runs, one negative (DNase-free water) and positive (H. pylori ATCC® 26695™) control was included.
Primers and thermal conditions in genotyping of H. pylori clinical isolates.
| Region amplified | Primer sequence(5′ to 3′) | Size ofamplicon | PCR cycles |
|---|
| glmM-F | AAGCTTTTAGGGGTGTTAGGGGTTT | 294 bp | Initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 45 sec, annealing at 50 °C for 45 sec and extension at 72 °C for 35 sec. The final extension was at 72 °C for 10 min. |
| glmM-R | AAGCTTACTTTCTAACACTAACGC |
| oipA-F | GTTTTTGATGCATGGGATTT | 401 bp | 5 min predenaturation at 95 °C, followed by 35 cycles of 35 sec at 95 °C, 35 sec at 53 °C and 35 sec at 72 °C |
| oipA-R | GTGCATCTCTTATGGCTTT |
| sabA-F | CCGCTAGTGTCCAGGGTAAC | 364 bp | 35 sec at 95 °C, 35 sec at 50 °C and 35 sec at 72 °C |
| sabA-R | CACCGCGATTTGCGTTGGTA |
| babA2-F | AATCCAAAAAGGAGAAAAATATGAAA | 832 bp | 35 sec at 95 °C, 35 sec at 55 °C and 45 sec at 72 °C.The final extension was at 72 °C for 5 min. |
| babA2-R | TGTTAGTGATTTCGGTGTAGGACA |
F: Forward primer, R: Reverse primer
DNA Sequence Analysis
The oipA, SabA and babA2 amplicons (one sample from each gene) were sequenced to verify that they represented the studied virulence genes. The sequencing was performed by Bioneer Company (Munpyeongseoro, Daedeok-gu, Daejeon, South Korea). The resulting sequences were edited, aligned and analyzed using the CLC Sequence Viewer (version 6.4; CLC Bio Co., Aarhus, Denmark).
Statistical Analysis
The Chi-square (χ2) test was used to analyze significant differences between the studied virulence genes with the clinical outcome. The data were analyzed using SPSS (version 21.0; IBM Co., Armonk, NY, USA) software. The results of demographic and clinical manifestations presented as descriptive statistics in terms of relative frequency. Differences were considered significant when p-value was less than 0.05 for data analysis.
Results
H. pylori Status and Clinical Outcomes
The isolation rate of H. pylori was 31.4%. The H. pylori-specific glmM (ureC) gene (294 bp) was confirmed in all isolates. The study population comprised of 50 men and 50 women with an average age of 42.9, SD=15.32 and 40.3, SD=13.40, respectively (range 18 to 75 years). A statistically significant difference between patients gender with four studied disease groups (p<0.014) and isolates distribution with disease status (p<0.001) was determined [20].
Distribution and Combined Presence of Virulence-Associated oipA, SabA and babA2 Genotypes in Different Disease Groups
The primers used for oipA and SabA genes amplified the corresponding genes in all isolates (100%). Nevertheless, the 832-bp amplicon indicating the presence of babA2 genotype [Table/Fig-2] was only detected in 22% (22 of 100) of isolates and the remaining isolates (78%) failed to obtain the babA2 gene amplification [Table/Fig-3]. There was no significant difference between each of the studied genes with clinical outcome (p>0.05). Moreover, no statistically significant difference was determined between oipA, SabA and babA2 genes and patient’s age or gender.
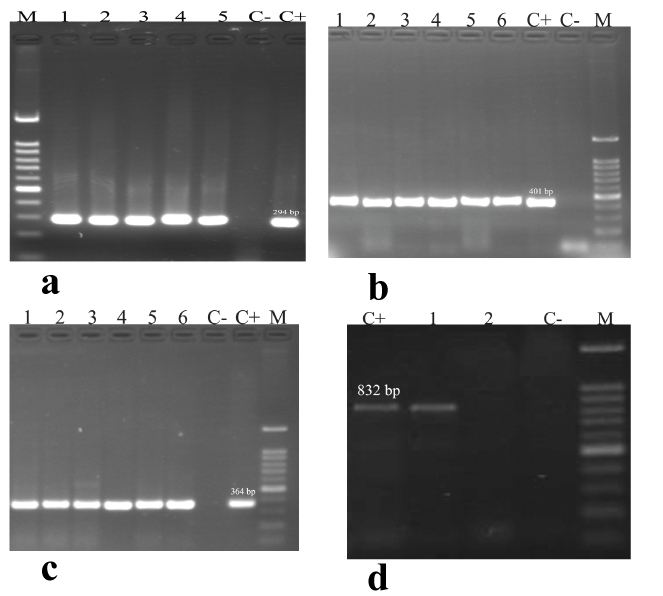
Agarose gel electrophoresis of glmM, oipA, sabA and babA2 genes in H. pylori isolates. A: M=DNA ladder (100 bp), C+ is positive control (H. pylori ATCC 26695, glmM= 294 bp), C- is negative control (DNase-free water), lanes 1-5 (positive samples). B: M=DNA ladder (100 bp), lanes 1-6 (positive samples of oipA=401 bp). C: M=DNA ladder (100 bp), lanes 1-6 (positive samples of sabA =364 bp). D: M=DNA ladder (100 bp), lane 1 (positive sample of babA2 =832 bp) and lane 2 (negative sample).
Distribution of H. pylori virulence genes and combined genotypes within disease groups.
| Virulence gene | GaNo. (%) | GUNo. (%) | DUNo. (%) | NUDNo. (%) | TotalNo. (%) |
|---|
| oipA | 63 (100) | 15 (100) | 13 (100) | 9 (100) | 100 (100) |
| sabA | 63 (100) | 15 (100) | 13 (100) | 9 (100) | 100 (100) |
| babA2 | 13 (20.6) | 4 (26.7) | 3 (23.1) | 2 (22.2) | 22 (22) |
| Genotype |
| oipA+/sabA+/babA2- | 51 (65.3) | 10 (12.8) | 11 (14) | 6 (7.6) | 78 (78) |
| oipA+/sabA+/babA2+ | 12 (54.5) | 5 (22.7) | 2 (9) | 3 (13.6) | 22 (22) |
*Ga: Gastritis, GU: Gastric ulcer, DU: Duodenal ulcer, NUD: Non-ulcer dyspepsia
Based on analysis of the oipA, SabA and babA2 genes (positive and negative), two different genotypic combinations were determined [Table/Fig-3]. The most prevalent genotype was oipA +/SabA +/ babA2- (78%). The oipA +/SabA +/ babA2- genotype was significantly associated only with Ga cases (p<0.001).
Discussion
Three factors including host’s genetics, environmental elements such as diet and H. pylori virulence genes are related to the severity of clinical manifestations [11, 23]. Among virulence genes, adhesins are very crucial for bacterial colonization and pathogenesis [6,15]. In recent years, there has been a great interest about the role of H. pylori OMPs, such as oipA, SabA and babA2 in developing of digestive diseases. Therefore, the presence and relationship of these virulence genes with clinical outcome needs to be further investigated. In the present study, H. pylori were isolated from 31.4% of patients. This result is similar to Abdollahi H et al., report from Iran [24], which implies the improvement of hygiene and the socio-economic conditions. Moreover, all studied isolates possessed both oipA and SabA genes, while the babA2 genotype has been only detected in 22% of isolates. However, no significant difference between each of studied genes with patients’ gender or age was determined.
Mansour KB et al., detected the oipA genotype in 90.8% of H. pylori clinical isolates from Tunisia [21]. In another study from Bulgaria, all 69 clinical isolates were detected positive for oipA and there was a strong association between oipA and peptic ulcer status [25]. In the survey of Yamaoka Y et al., [5], oipA positive isolates from the US and Colombian patients were significantly recognized in duodenal ulcers (88%) and gastric cancer (89%) subjects. Several studies in Iran showed different ranges for prevalence of oipA from 40.8% to 95.9% [26-30]. Nevertheless, in none of these studies, except two of them [29,30] an association has been found between the genes and clinical outcome. Our result is similar to the obtained data by Lehours P et al., [12], who found that all H. pylori isolates were oipA positive. However, they did not find any correlation between oipA gene with different gastroduodenal diseases. Our finding is also in accordance with reports from some other studies [17, 28, 31], which reflects the importance of geographic differences in H. pylori virulence factors distribution [25]. Nevertheless, some works have shown an association between OipA with peptic ulcers and gastric malignancies [5,6,16].
The role of SabA in gastroduodenal disorders is progressively apparent. Due to absence of sLeX in the healthy gastric mucosa, it was proposed that the SabA is related to the H. pylori chronic persistence [9]. Therefore, it is suggested that SabA is related to severe clinical outcomes, such as gastric cancer [16]. In two studies from Japan, a high frequency of SabA gene among H. pylori clinical isolates (with frequencies of 91. 3% and 81%, respectively) has been reported [1, 32]. In the latter one, there was no significant correlation between the SabA genotype with gastric diseases. Conversely, in other report from Japan [33], the rate of SabA was found 47.7% and it was also suggested that the presence of SabA gene might not correlated with Ga. In some previous studies, no association between SabA and clinical manifestations was reported [12,17,34,35]. Yamaoka Y and co-workers [5] assessed 200 H. pylori isolates from the US and Colombian patients, in which the SabA positive status was associated with gastric cancer and Ga. In two different studies from Iran, the prevalence of SabA have reported 83.3%-100% and 83.6%, respectively [36,37], with no association with the severity of clinical manifestations. In our study, the SabA genotype showed a high frequency (100%), which is somewhat similar to Shao L et al., investigation [1]. In the study of Yanai A et al., [32], inspite of the positivity of the most of H. pylori isolates for functional SabA; but, this gene was not as a marker for gastroduodenal disorders, which is consistent with our findings. This discrepancy between our results and other reports, underlines that SabA genetically varies among different strains and geographical areas [1] and also may not be present in all H. pylori isolates [16]. It is necessary to note that patient selection to establish an association between H. pylori virulence genes and clinical outcome is very important, because the studied groups should be sufficiently large and heterogeneous [21].
It has been mentioned that H. pyloribabA2 positive isolates are associated with increased risk of acute gastritis, duodenal ulcers and gastric cancer and that the babA2 detection in clinical isolates by PCR does not necessarily correlates with its adhesive properties and vice versa [12,38]. In two studies conducted among Colombian patients, it was found that babA2 gene was more related to duodenal ulcers and gastric cancer [5,22], which is not in agreement with our observation. In contrast, in a research from Turkey [31], no significant difference was found between babA and other virulence genes with Ga and ulcer. Con SA et al., [39] reported a high prevalence of babA2, 73.7% and 96.8% in Costa Rican and Japanese isolates, respectively. In Brazil, 46.15% strains were babA2 positive and a strong association has been observed between the presence of babA2 and duodenal ulcers as well as gastric carcinoma [40]. Yu J et al., [41] have shown that around 80% of studied patients were infected with babA2 positive strains. The babA2 prevalence among South-American H. pylori isolates was reported between 46-82.3% [38]. In Iran, these fluctuations vary from 49.5% to 96.7% [29,30,35,42,43], with no observed correlation between babA2 and peptic ulcer disease or gastric cancer. Our result (babA2=22%) is in agreement with Paniagua GL and co-workers survey [38]; however, it is considerably lesser than other studies from Iran. Moreover, no significant association was seen between the presence of babA2 genotype and clinical manifestations. This result is supported by some other works [12,34]. Notably, frequency of babA2 in our isolates was low similar to data from Colombia and Cuba, countries with low incidence of gastric cancer [22,44] and in contrary with Japan, Costa Rica, Brazil and China where a high rate of gastric cancer have been found [39-41]. On the other hand, the most prevalent combined genotype in the present study was oipA+/ SabA+/babA2- (78%), which was significantly associated with Ga.
The results of the present study highlight the importance of oipA and SabA genes in H. pylori infection associated complications. Indeed, it seems that the most important point is low frequency of babA2 genotypes in our isolates which is in contrast to other regions of Iran.
Limitation
There are some limitations in our study such as sample size, which was relatively small. Lack of gastric cancer cases among our studied samples was another limitation for our study; thereby we could not evaluate the studied genes among H. pylori isolates from those patients. We also did not evaluate the oipA functional status among our isolates.
Conclusion
Our study showed a considerably low frequency of babA2 genotype among H. pylori isolates examined from Southwest of Iran. This result is different from other Iranian reports; however, in agreement with countries with a low rate of gastric malignancy. Therefore, with respect to heterogeneity of H. pylori strains and diverse results from different geographic regions about above mentioned genotypes, further investigations are required to assess the role of these genes in H. pylori pathogenesis and their association with clinical outcomes.
[1]. Shao L, Takeda H, Fukui T, Mabe K, Han J, Kawata S, Genetic diversity of the Helicobacter pylori sialic acid-binding adhesin (sabA) geneBiosci Trends 2010 4(5):249-53. [Google Scholar]
[2]. Haddadi MH, Bazargani A, Khashei R, Fattahi MR, Bagheri Lankarani K, Moini M, Different distribution of Helicobacter pylori EPIYA- cagA motifs and dupA genes in the upper gastrointestinal diseases and correlation with clinical outcomes in Iranian patientsGastroenterol Hepatol Bed Bench 2015 8:S37-46. [Google Scholar]
[3]. Espinoza MG, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG, Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCRJ Clin Microbiol 2011 49(4):1650-52. [Google Scholar]
[4]. Singh V, Mishra S, Rao GR, Jain AK, Dixit VK, Gulati AK, Evaluation of nested PCR in detection of Helicobacter pylori targeting a highly conserved gene: HSP60Helicobacter 2008 13:30-34. [Google Scholar]
[5]. Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, Helicobacter pylori outer membrane proteins and gastroduodenal diseaseGut 2006 55(6):775-81. [Google Scholar]
[6]. Kalali B, Mejías-Luque R, Javaheri A, Gerhard M, H. pylori virulence factors: influence on immune system and pathologyMediators Inflamm 2014 4263092014 [Google Scholar]
[7]. Odenbreit S, Swoboda K, Barwig I, Ruhl S, Borén T, Koletzko S, Outer membrane protein expression profile in Helicobacter pylori clinical isolatesInfect Immun 2009 77(9):3782-90. [Google Scholar]
[8]. Yakoob J, Abbas Z, Khan R, Salim SA, Awan S, Abrar A, Helicobacter pylori outer membrane protein Q allele distribution is associated with distinct pathologies in PakistanInfect Genet Evol 2016 37:57-62. [Google Scholar]
[9]. Posselt G, Backert S, Wessler S, The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces amulti-step process in pathogenesisCell Commun Signal 2013 11:77 [Google Scholar]
[10]. Boyanova L, Yordanov D, Gergova G, Markovska R, Mitov I, Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristicsAntonie Van Leeuwenhoek 2010 98(3):343-50. [Google Scholar]
[11]. Ortiz-Princz D, Daoud G, Salgado-Sabel A, Cavazza ME, Helicobacter pylori infection in children: should it be carefully assessed?Eur Rev Med Pharmacol Sci 2016 20(9):1798-813. [Google Scholar]
[12]. Lehours P, Ménard A, Dupouy S, Bergey B, Richy F, Zerbib F, Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphomaInfect Immun 2004 72(2):880-88. [Google Scholar]
[13]. Yamaoka Y, Pathogenesis of Helicobacter pylori-related gastroduodenal diseases from molecular epidemiological studiesGastroenterol Res Pract 2012 :371503 [Google Scholar]
[14]. Backert S, Clyne M, Tegtmeyer N, Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pyloriCell Commun Signal 2011 9:28 [Google Scholar]
[15]. Skindersoe ME, Rasmussen L, Andersen LP, Krogfelt KA, A novel assay for easy and rapid quantification of Helicobacter pylori adhesionHelicobacter 2015 (3):199-205. [Google Scholar]
[16]. Yamaoka Y, Increasing evidence of the role of Helicobacter pyloriSabA in the pathogenesis of gastroduodenal diseaseJ Infect Dev Ctries 2008 2(3):174-81. [Google Scholar]
[17]. Chiarini A, Calà C, Bonura C, Gullo A, Giuliana G, Peralta S, Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, ItalyEur J Clin Microbiol Infect Dis 2009 28(5):437-46. [Google Scholar]
[18]. Colbeck JC, Hansen LM, Fong JM, Solnick JV, Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pyloriInfect Immun 2006 74(7):4375-78. [Google Scholar]
[19]. Zhang J, Qian J, Zhang X, Zou Q, Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pyloriMol Biol Rep 2014 41(12):7807-14. [Google Scholar]
[20]. Khashei R, Dara M, Bazargani A, Bagheri Lankarani K, Taghavi A, Moeini M, High rate of A2142G point mutation associated with clarithromycin resistance among Iranian Helicobacter pylori clinical isolatesAPMIS 2016 124:787-93. [Google Scholar]
[21]. Mansour KB, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patientsAnn Clin Microbiol Antimicrob 2010 9:10 [Google Scholar]
[22]. Arévalo-Galvis A, Trespalacios-Rangell AA, Otero W, Mercado-Reyes MM, Poutou-Piñales RA, Prevalence of cagA,vacA, babA2 and iceA genes in H.pylori strains isolated from Colombian patients with functional dyspepsiaPol J Microbiol 2012 61(1):33-40. [Google Scholar]
[23]. Shiota S, Suzuki R, Yamaoka Y, The significance of virulence factors in Helicobacter pyloriJ Dig Dis 2013 14(7):341-49. [Google Scholar]
[24]. Abdollahi H, Savari M, Zahedi MJ, Moghadam SD, Abasi MH, Detection of A2142C, A2142G, and A2143G mutations in 23s rRNA gene conferring resistance to clarithromycin among Helicobacter pylori isolates in Kerman, IranIran J Med Sci 2011 36(2):104-10. [Google Scholar]
[25]. Markovska R, Boyanova L, Yordanov D, Gergova G, Mitov I, Helicobacter pylorioipA genetic diversity and its associations with both disease and cagA, vacA s, m, and i alleles among Bulgarian patientsDiagn Microbiol Infect Dis 2011 71(4):335-40. [Google Scholar]
[26]. Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, IranJ Gastroenterol Hepatol 2009 24(8):1380-86. [Google Scholar]
[27]. Aghdam SM, Sardari Z, Safaralizadeh R, Bonyadi M, Abdolmohammadi R, Moghadam MS, Investigation of association between oipA and iceA1/iceA2 genotypes of Helicobacter pylori and gastric cancer in IranAsian Pac J Cancer Prev 2014 15(19):8295-8299. [Google Scholar]
[28]. Souod N, Sarshar M, Dabiri H, Momtaz H, Kargar M, Mohammadzadeh A, The study of the oipA and dupA genes in Helicobacter pylori strains and their relationship with different gastroduodenal diseasesGastroenterol Hepatol Bed Bench 2015 8(Suppl1):S47-S53. [Google Scholar]
[29]. Sedaghat H, Moniri R, Jamali R, Arj A, Razavi Zadeh M, Moosavi SG, Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseasesIran J Microbiol 2014 6(1):14-21. [Google Scholar]
[30]. Salimzadeh L, Bagheri N, Zamanzad B, Azadegan-Dehkordi F, Rahimian G, Hashemzadeh-Chaleshtori M, Frequency of virulence factors in Helicobacterpylori-infected patients with gastritisMicrob Pathog 2015 80:67-72. [Google Scholar]
[31]. Oktem-Okullu S, Tiftikci A, Saruc M, Cicek B, Vardareli E, Tozun N, Multiplex-PCR-based screening and computational modeling of virulence factors and T-Cell mediated immunity in Helicobacter pylori infections for accurate clinical diagnosisPLoS One 2015 10(8):e0136212 [Google Scholar]
[32]. Yanai A, Maeda S, Hikiba Y, Shibata W, Ohmae T, Hirata Y, Clinical relevance of Helicobacter pylorisabA genotype in Japanese clinical isolatesJ Gastroenterol Hepatol 2007 22(12):2228-32. [Google Scholar]
[33]. Nakasato F, Shimoyama T, Yoshimura T, Mikami T, Munakata A, Fukuda S, Infection of sabA-positive H. pylori does not induce anti-Lewis X antibody in hostHepatogastroenterol 2008 55(84):1122-25. [Google Scholar]
[34]. Gasiorowska J, Parzecka M, Szaflarska-Poplawska A, Gorzkiewicz M, Grzybowski T, Polymorphism of Helicobacter pylori and the presence of genes babA2 and sabA and endoscopic and histopathological changes in patients infected with Heicobacter pyloriPol Merkur Lekarski 2013 35(208):191-95. [Google Scholar]
[35]. Yadegar A, Mobarez AM, Alebouyeh M, Mirzaei T, Kwok T, Zali MR, Clinical relevance of cagL gene and virulence genotypes with disease outcomes in a Helicobacter pylori infected population from IranWorld J Microbiol Biotechnol 2014 30(9):2481-90. [Google Scholar]
[36]. Pakbaz Z, Shirazi MH, Ranjbar R, Pourmand MR, Khalifeh Gholi M, Aliramezani A, Frequency of sabA gene in Helicobacter pylori strains isolated from patients in Tehran, IranIran Red Crescent Med J 2013 15(9):767-70. [Google Scholar]
[37]. Yadegar A, Alebouyeh M, Zali MR, Analysis of the intactness of Helicobacter pylori cag pathogenicity island in Iranian strains by a new PCR-based strategy and its relationship with virulence genotypes and EPIYA motifsInfect Genet Evol 2015 35:19-26. [Google Scholar]
[38]. Paniagua GL, Monroy E, Rodríguez R, Arroniz S, Rodríguez C, Cortés JL, Frequency of vacA, cagA and babA2 virulence markers in Helicobacter pylori strains isolated from Mexican patients with chronic gastritisAnn Clin Microbiol Antimicrob 2009 8:14 [Google Scholar]
[39]. Con SA, Takeuchi H, Nishioka M, Morimoto N, Sugiura T, Yasuda N, Clinical relevance of Helicobacter pylori babA2 and babA2/B in Costa Rica and JapanWorld J Gastroenterol 2010 16(4):474-78. [Google Scholar]
[40]. Oliveira AG, Santos A, Guerra JB, Rocha GA, Rocha AM, Oliveira CA, babA2- and cagA-positive Helicobacter pylori strains are associated with duodenal ulcer and gastric carcinoma in BrazilJ Clin Microbiol 2003 41(8):3964-66. [Google Scholar]
[41]. Yu J, Leung WK, Go MY, Chan MC, To KF, Ng EK, Relationship between Helicobacter pylori babA2 status with gastric epithelial cell turnover and premalignant gastric lesionsGut 2002 51(4):480-84. [Google Scholar]
[42]. Shahi H, Reiisi S, Bahreini R, Bagheri N, Salimzadeh L, Shirzad H, Association between Helicobacter pylori cagA, babA2 virulence factors and gastric mucosal Interleukin-33 mRNA expression and clinical outcomes in dyspeptic patientsInt J Mol Cell Med 2015 4(4):227-34. [Google Scholar]
[43]. Vaziri F, Najar Peerayeh S, Alebouyeh M, Mirzaei T, Yamaoka Y, Molaei M, Diversity of Helicobacter pylori genotypes in Iranian patients with different gastroduodenal disordersWorld J Gastroenterol 2013 19(34):5685-92. [Google Scholar]
[44]. Torres LE, Melián K, Moreno A, Alonso J, Sabatier CA, Hernández M, Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolatesWorld J Gastroenterol 2009 15(2):204-10. [Google Scholar]