Coagulase-Negative Staphylococcus (CoNS) is considered as an important agent of nosocomial or healthcare-associated infections among immunosuppressed patients and elderly individuals [1,2]. In the past decades, CoNS was considered as a commensal bacterium and normal bacterial flora of healthy human skin, but currently, these organisms are considered as common cause of human diseases [3]. CoNS has also been found the third most common causative agent of nosocomial infections. Among CoNS, Staphylococcus epidermidis is an important pathogen in neonatal sepsis and patients who use medical equipments such as urinary catheters and other implanted devices [4]. Approximately 75%-90% of these bacteria, circulating in hospitals are resistance to Methicillin [5-7]. The resistance of S. epidermidis to methicillin is encoded by the expression of Penicillin Binding Protein (PBP2a) with a low affinity for this antibiotic. PBP2a is encoded by the mecA gene, which is located on a mobile genetic element and it is called the SCCmec [8].
Materials and Methods
Study Design and Population
This cross-sectional study was carried out from March 2014 to January 2015 in Al-Zahra hospital, affiliated to Isfahan University of Medical Sciences, Isfahan, Iran. All the parts of study were supervised and approved by the Ethical Committee of Isfahan University of Medical Sciences (code 293253).
Bacterial Isolation and Identification
One hundred and thirty S. epidermidis isolates were collected from clinical samples of patients who referred to Isfahan’s AL-Zahra hospital (Iran). The isolates were recovered from wounds (n=30), blood (n=18), urine (n=60), sputum (n=7), Bronchoalveolar Lavage (BAL) (n=5), and abscesses (n=10). Thereafter, all isolates were identified as S. epidermidis using different conventional methods including Gram staining, catalase test, plasma coagulase test, and final identification of S. epidermidis was performed by the ability of urease production and carbohydrate fermentation [10]. All strains were stored at -70°C in Trypticase Soy Broth (TSB), with 20% glycerol.
Antimicrobial Susceptibility Assay
Antimicrobial susceptibility tests of S. epidermidis isolates were performed by the disc diffusion method on Mueller– Hinton agar (Himedia, India) according to the guidelines of Clinical and Laboratory Standards Institute (CLSI) for gentamycin (10 μg), oxacillin (1 μg), tetracycline (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), Co-Trimoxazole (SXT- 1.25/23.75 μg), rifampin (5 μg), and vancomycin antibiotic discs (MAST, UK) [20]. S. aureus ATCC® 25923™, which is a methicillin-sensitive S. aureus, was used as the control strain in antibacterial susceptibility testing. All the strains were screened for methicillin resistance based on resistance to cefoxitin (30 μg) discs (MAST, UK) by the disc diffusion method according to the CLSI 2013 guidelines.
DNA Extraction and PCR Assay
Bacterial genomic DNA of suspected isolates were extracted directly from 24 hours colonies by boiling a dense suspension in sterile distilled water for 10 minutes and centrifuged for two minutes at 13,000×g [21]. The genomic DNA was used as a template for typing of SCCmec, detection of mecA and the PVL cytotoxin gene. The PCR for amplification of mecA gene was performed as described previously [22].
Detection of the PVL Gene
PCR was performed to determine the presence of the PVL cytotoxin gene as previously described by Azimian A et al., [22].
SCCmec Typing by Multiplex-PCR
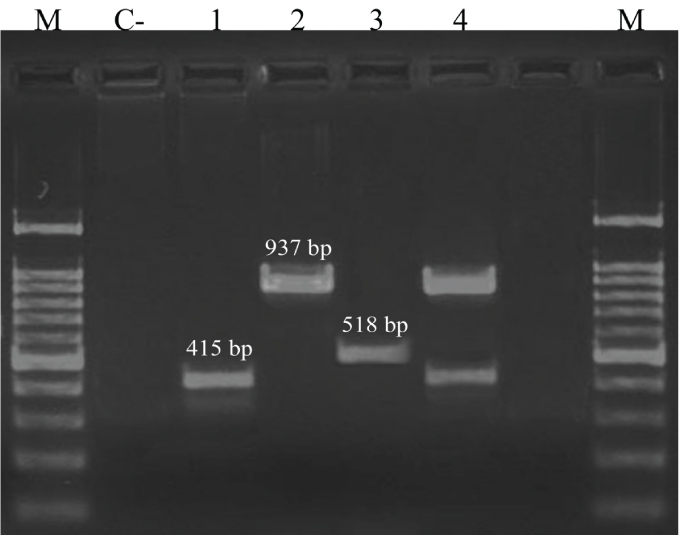
The SCCmec type was determined in a multiplex PCR which included four pairs of primers for the SCCmec types I- IV. PCR was performed in a mixture of 30 μl volume containing 15 μl master mix, 0.5 μl of each of α3R1, βF1, ccrCF and ccrCR primers (10 pmol/μl), 0.3 μl of each of 1272F1, 1272R1, 5RmecAF, 5R431R primers (10 pmol/μl) and 2 μl template DNA. The final volume was adjusted to 30 μl by adding 9.8 μl sterile ultrapure water and was placed on thermal cycler [23]. Amplification protocol consisted of five minutes initial denaturation at 93°C, followed by 30 cycles of denaturation (93°C/45 seconds), annealing (55°C/30 seconds), and extension (72°C/90 seconds), and an additional post-amplification extension step at 72°C for five minutes. The primers used for the PCR are listed in [Table/Fig-1]. The following S. aureus reference strains were used as controls: COL; N315; 85/2082; and JCSC/4469 for the SCCmec types I, II, III, and IV, respectively [Table/Fig-2].
List of primers used in this study.
| Target | Primer | Sequence | Product size (bp) | Reference |
|---|
| SCCmec | β F1 | ATTGCCTTGATAATAGCCYTCT | 937 | [23] |
| α3 R1 | TAAAGGCATCAATGCACAAACACT |
| ccrC F | CGTCTATTACAAGATGTTAAGGATAAT | 518 | [23] |
| ccrC R | CCTTTATAGACTGGATTATTCAAAATAT |
| 1272 F | GCCACTCATAACATATGGAA | 415 | [23] |
| 1272 R | CATCCGAGTGAAACCCAAA |
| 5RmecAF | TATACCAAACCCGACAACTAC | 359 | [23] |
| 5R431R | CGGCTACAGTGATAACATCC |
| PVL | PVL F | GGAAACATTTATTCTGGCTATAC | 502 | [22] |
| PVL R | CTGGATTGAAGTTACCTCTGG |
| mecA | F | AGAAGATGGTATGTGGAAGTTAG | 583 | [23] |
| R | ATGTATGTGCGATTGTATTGC |
*F: Forward, R: Reverse
C-: negative control (distilled water); lane 1: SCCmec type I (COL); lane 2: SCCmec type II (N315); lane 3: SCCmec type 3 (85/2082); lane 4: SCCmec type IV (JCSC/4469); M: 100 bp ladder.
Statistical Analysis
Data were entered and analysed by Statistical Package for the Social Sciences (SPSS) software version 20.0. The description of the results was carried out by frequencies. For categorical variables different groups were compared using the Chi-square test or Fisher-exact test. All probabilities were two-tailed and a p-values < 0.05 was considered to be statistically significant.
Results
Frequency of MRSE
Out of total 130 S. epidermidis isolates, the frequency of MRSE was 52.3% (n=68) and 53.8% (n=70) according to the disc diffusion and presence of mecA gene, respectively. Phenotypic (Cefoxitin disc) and molecular (mecA detection) based MRSE screening methods showed different results [Table/Fig-3]. Specimen wise distribution showed that MRSE was most frequent in urine (53%), followed by blood (11.4%), sputum (5.7%), wound (25.6%) and BAL (4.3%). On the other hand, the frequency of MRSE was found to be 42.7% and 57.3% for female and male respectively.
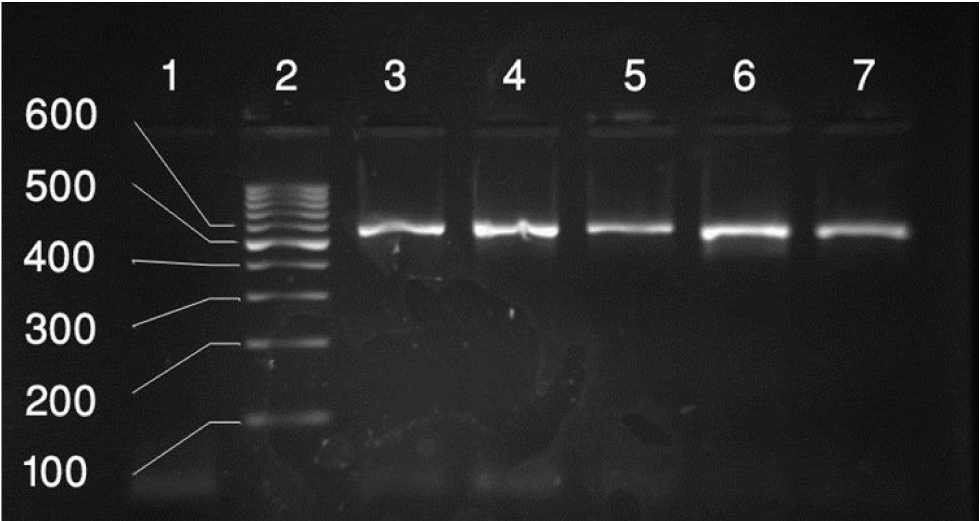
PCR amplification of the mecA genes. Lane 1: Negative Control, Lane 2: 100 bp-3k b ladder, lane 3: positive control for mecA genes (583bp), lane 4-7: positive results for mecA genes.
Antibiotic Susceptibility Pattern of S. epidermidis Isolates
The antibiotic susceptibility showed that all MRSE isolates were 100% sensitive to vancomycin. The overall resistance rates were high with ciprofloxacin (81.4%) and co-trimoxazole (64%), while the lowest level of antibiotics resistance were against gentamycin (37.1%). [Table/Fig-4] shows the distribution of antibiotic resistance among MRSE isolates based on SCCmec type.
Distribution of antibiotic resistance among MRSE isolates by SCCmec types.
| Antibiotic | SCCmec I (n=30) | SCCmec II (n=8) | SCCmec IV (n=24) | Untypeable (n=8) | Total (n=70) |
|---|
| Gentamycin | 11 | 1 | 12 | 2 | 26 |
| Oxacillin | 24 | 5 | 24 | 6 | 59 |
| Tetracycline | 23 | 3 | 15 | 3 | 44 |
| Ciprofloxacin | 26 | 6 | 19 | 6 | 57 |
| Clindamycin | 15 | 4 | 15 | 4 | 38 |
| Co-trimoxazole | 19 | 5 | 15 | 6 | 45 |
| Rifampin | 14 | 3 | 15 | 0 | 32 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 |
Characterization of SCCmec Type and PVL Gene
Results from PCR analysis shown that 12 (17%) of MRSE isolates carried the PVL gene [Table/Fig-5]. In addition, of the 70 MRSE isolates, 43% (30/70) were SCCmec type I; 11.4% (8/70) were type II; 34.2% (24/70) were type IV and 11.4 (8/70) of the MRSE isolates could not be typed, whereas, none of the isolates were identified as SCCmec type III [Table/Fig-6]. The SCCmec type I of MRSE isolates were significantly (p<0.05) recovered as the major type from 19 (63.3%) urine followed by 5 (16.7%) from wound, 3 (10%) from blood and 3 (10%) from sputum, while the SCCmec type II isolates were 4 (50%) from sputum, 3 (37.5%) from urine, 1 (12.5%) from wound and the SCCmec type IV isolates were mostly 13 (54.1%) isolated from the urine samples, 8 (33.4%) from blood and 3 (12.5%) from sputum. Also, the majority of PVL-positive isolates were SCCmec type I (50%).
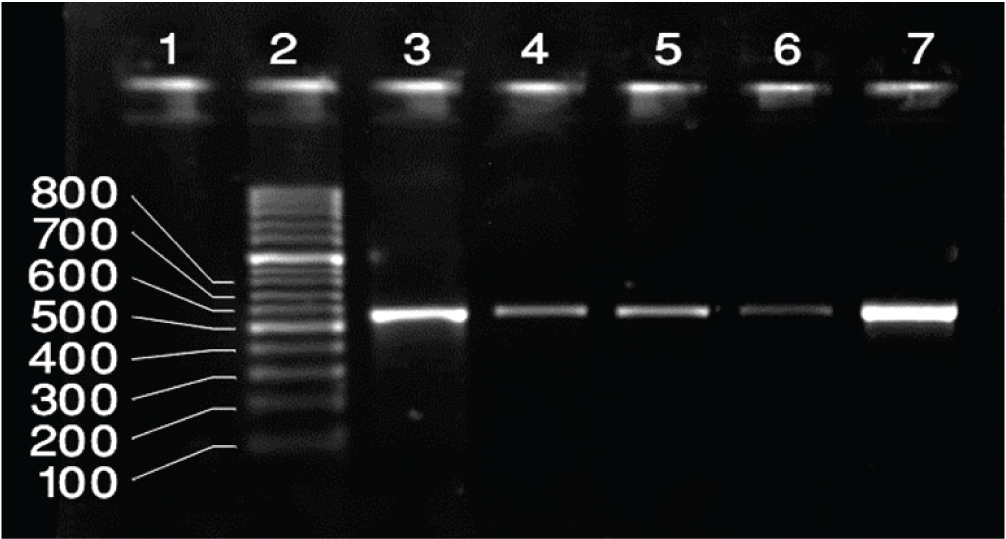
PCR amplification of the PVL genes. Lane 1: Negative Control, Lane 2 100 bp-3kb ladder, lane 3: Positive control for PVL genes (502bp), Lane 4-7: Positive results for PVL genes.
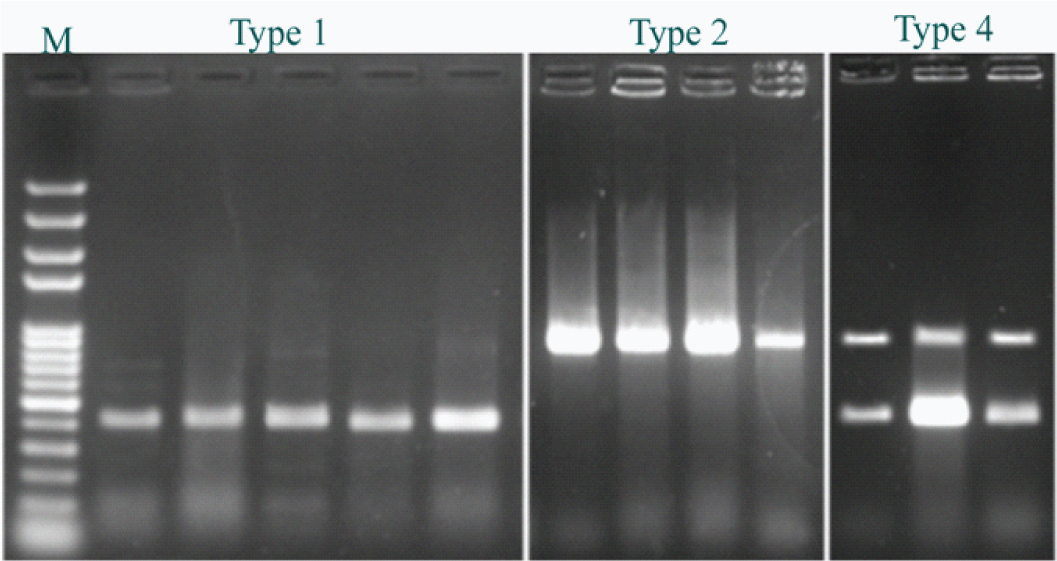
Positive results for SCCmec types I (415bp), II (937bp), IV (937/415 bp).
Discussion
S. epidermidis, especially MRSE, is one of the major causes of serious infections such as catheter-related blood stream infections [24]. Its various virulence factors and high rate of methicillin resistance among S. epidermidis, cause various problems for patients and healthcare providers during treatment [1]. To investigate outbreaks in nosocomial infections, determination of the source is very important that molecular typing is a valuable tool for this purpose [25].
The purpose of this study was to investigate SCCmec types, PVL gene and determining antimicrobial resistance patterns of clinical MRSE isolates collected from patient in Isfahan. Antibiotic resistance of MRSE to ciprofloxacin, cefoxitin, clindamycin, tetracycline, gentamycin, cotrimoxazole, rifampin and vancomycin using disk diffusion method was investigated. Our results showed that the prevalence of methicillin resistance among the S. epidermidis isolates was found to be 70 (53.8%) which is in contrast with Namvar AE et al., with 83.4% [23]. This resistance rate was lower than those found in the rest of Iran [23,26]. In a similar study carried out by Du X et al., prevalence of MRSE isolates was 86.7% among clinical isolates which are in contrast with the current study [27]. However, the prevalence of MRSE varied widely among different population and regions representing different health policies and other factors involved.
S. epidermidis isolates were resistant to many common antibiotics in the clinic. In antimicrobial susceptibility pattern of isolates, ciprofloxacin (81.4%) and co-trimoxazole (64%) had the highest resistance, while resistance to rifampin and gentamycin was 45.7% and 37.1% respectively. Also, all the isolates were completely sensitive to vancomycin. Overall, the rate of antibiotic resistance in our results was lower than the previous studies which can be associated with the pattern of antibiotic use in medical wards [15,26]. In the present study, SCCmec typing was used for detection and epidemiological investigations of isolates. Our findings show that a total of 70 MRSE isolates, SCCmec type were found in 88.5% (62/70) isolates. In other words, among MRSE isolates, the most common SCCmec type was type I, with a frequency rate of 43% (30/70), the second most frequent SCCmec was type IV with a rate of 34.2% (24/70) followed by type II in 11.4% (8/70) of isolates, whereas 11.4% (8/70) of the MRSE isolates could not be typed. Consistent with our results, Mertens A et al., revealed that 39.5% of S. epidermidis isolates carry SCCmec type I [28]. Also, in a study in Brazil, Machado ABMP et al., in 2013 indicated that 27.9% of S. epidermidis isolates harboured SCCmec type I, these data was similar to our results. In addition, Machado ABMP et al., showed no SCCmec type II in their population which is different with our finding [13]. In a study by Namvar AE et al., characterization of SCCmec in S. epidermidis identified 2 (2.1%) isolates harboured SCCmec type I, and the rate of SCCmec type IV were 42 (43.8%) which is in contrast to the results found in our study [23]. Possible reasons for the wide range of SCCmec type might be due to differences in geographical regions, the study population, and detection methods [29,30]. Our result is similar to studies which have shown that non-typeable SCCmec among S. epidermidis isolate’s rate is higher than others [2,23]. This higher rate might be due to the presence of other SCCmec types [31,32]. Most of the SCCmec type I and IV isolates were resistant to all antibiotics excluding vancomycin, while in comparison to other types, SCCmec type II isolates were found to be more sensitive to gentamycin, tetracycline and rifampin. Generally, among all of SCCmec types, the highest antibiotic sensitivity of the isolates was against gentamycin, followed by rifampin. Our results showed, 17% (12/70) carried the PVL gene and these data indicated a high rate of PVL gene among S epidermidis. In comparison with the present study, Sani NAM et al., revealed 8% of the S. epidermidis isolate had the complete operon of staphylococcal toxin genes such as PVL [33]. Also, in a study in Korea, PCR results for PVL revealed 15 (71.4%) of the S. epidermidis isolates carried the PVL genes [34]. Although, several reports demonstrate low prevalence of PVL gene similar to our findings which is usually associated with cutaneous infections. While, PVL gene for CoNS was absent in a number of studies [35,36]. Increased prevalence of PVL gene and SCCmec types among S. epidermidis isolates is an alarming situation. Further investigations are needed to be done on the same to prevent any complications regarding the treatment in near future.
Limitation
There were several limitation to our study; first, detection of other types of SCCmec and their subtypes was not performed. Second, the patients participated in the study were from one hospital causing difficulty to generalize our results to other hospitals. Last, our sample size was not enough to interpret the characterization of SCCmec and distribution of MRSE in our region. MIC of vancomycin and other antimicrobial agents was not determined.
Conclusion
Screening and isolation of MRSE-positive patients is essential in order to control the transmission of MRSE in both community and hospitals. SCCmec typing is useful in differentiating both and thus can be used for early diagnosis of infections.